Mars - Molecule archive suite is broad framework for image processing, storage and reproducible analysis of single-molecule data.
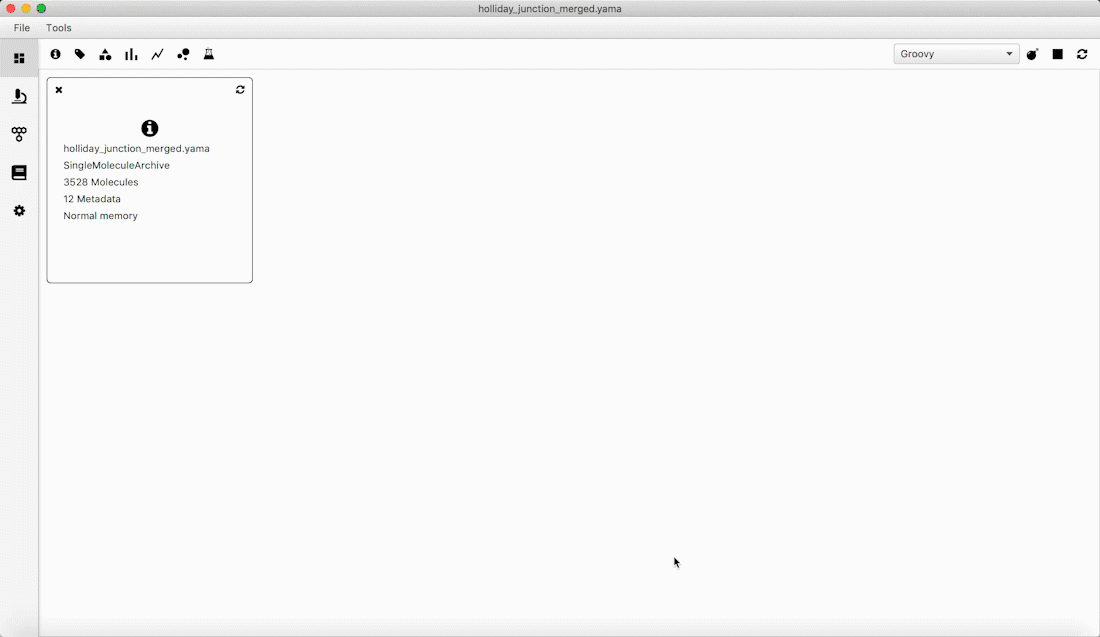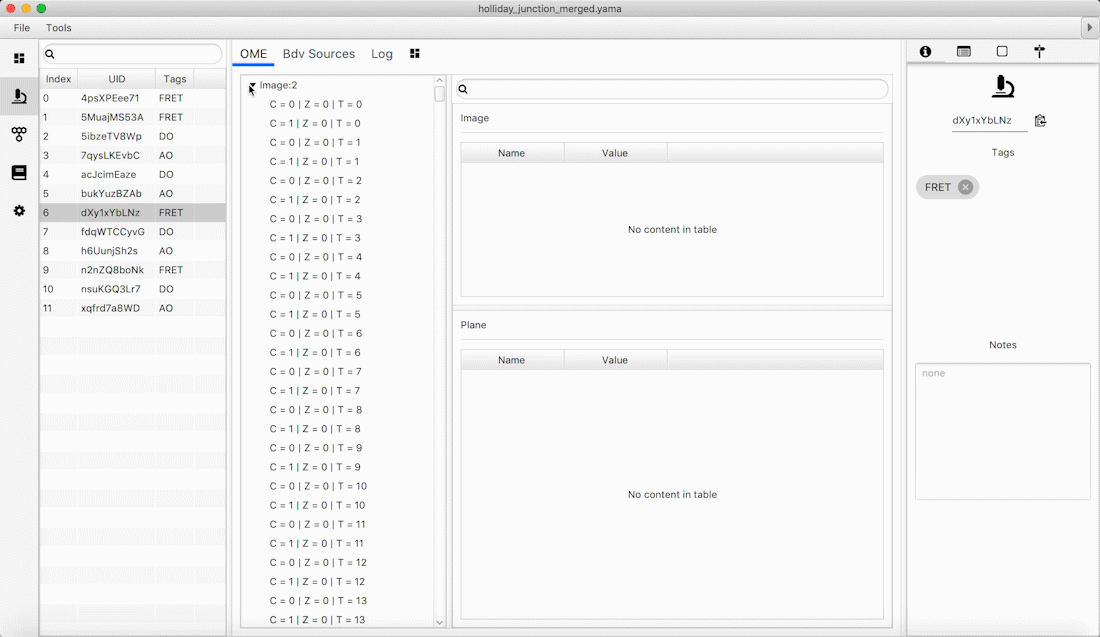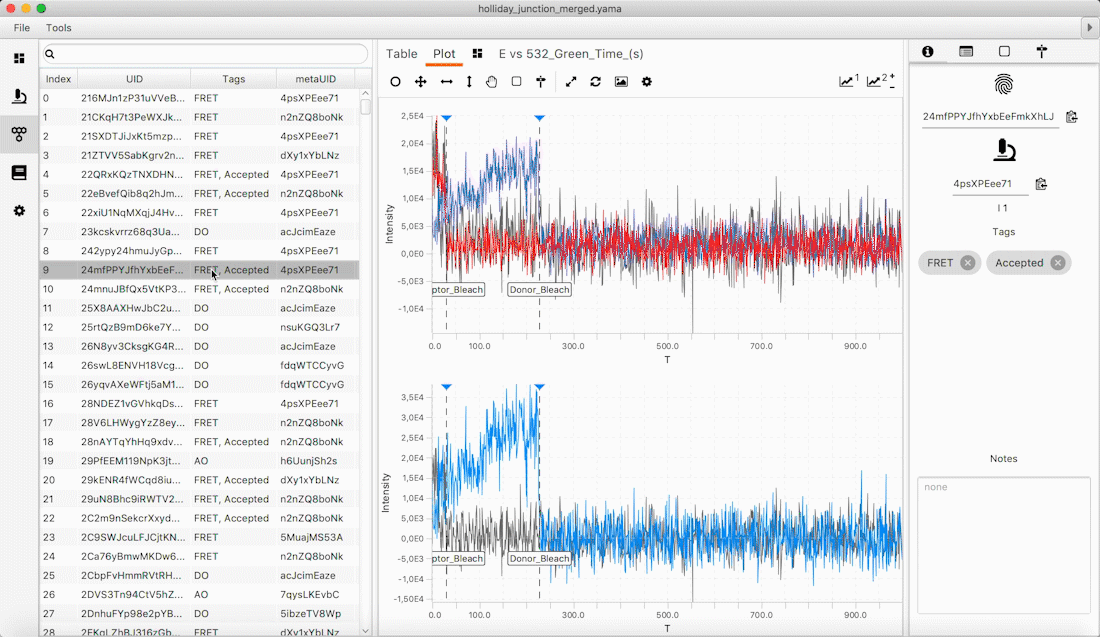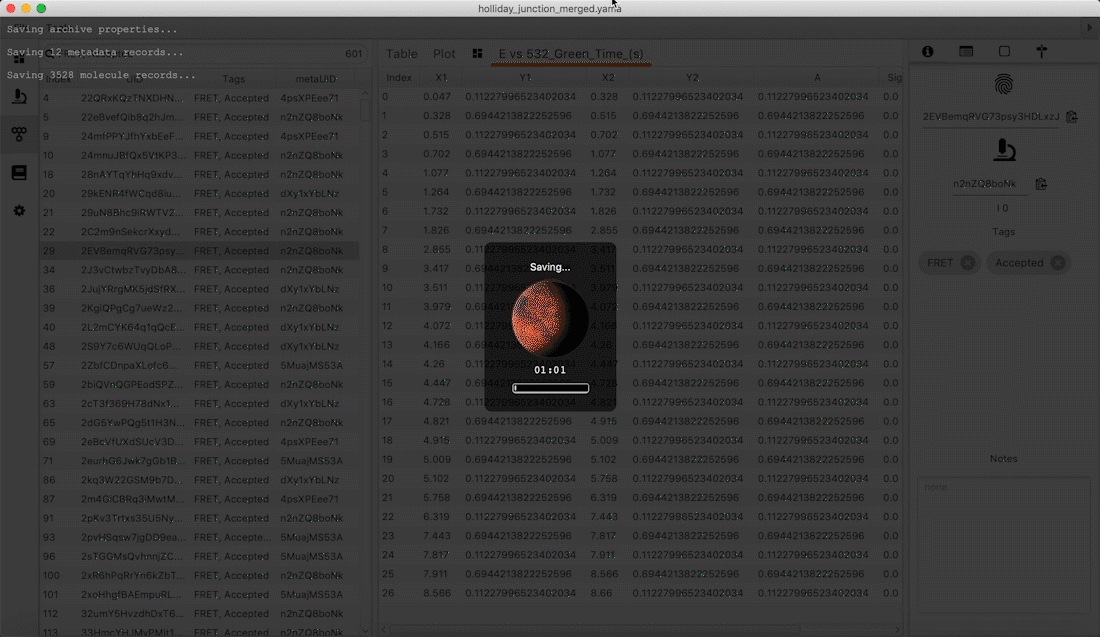
Mars provides a collection of ImageJ2/Fiji commands to find, fit, track and characterize single molecules. The algorithms provided are routinely used to analyze data arising from multicolor single molecule TIRF and DNA flow stretching experiments. Primary image analysis commands generate Molecule Archives which contain collections of individual biomolecule and image metadata records. Molecule Archives have a simple, yet powerful, API for fast and concise access to subsets of records based on arbitrary properties. During creation, all records are assigned universally unique IDs, which serve as the primary keys for retrieval and storage. This architecture allows for seamless virtual storage, merging, and multithreaded processing of very large datasets. Moreover, this framework allows for the same data structure to be used from initial processing all the way to the generation of final plots either in Fiji or in Jupyter notebooks using Python. Please cite our paper if you use Mars in your own work so we can continue development and support:
Nadia M Huisjes Thomas M Retzer Matthias J Scherr Rohit Agarwal Lional Rajappa Barbara Safaric Anita Minnen Karl E Duderstadt (2022) Mars, a molecule archive suite for reproducible analysis and reporting of single-molecule properties from bioimages eLife 11:e75899. https://doi.org/10.7554/eLife.75899
To get started with Mars, take a look at the tutorials, example gallery, and how to install guide, or read about the project’s goals. Comprehensive reference material can be found in the documentation section. If you have questions or problems, or would just like to tell us about your experience with Mars, please get in touch with us on the Scientific Community Image Forum using the mars tag.