About the Mars Project
For Reproducible Analysis of Single-Molecule Observations go to Mars
The Reproducibility Crisis
Reproducible analysis of bioimaging data is a major challenge slowing scientific progress. The increasing size of acquired datasets and the heterogeneity of data storage formats impedes sharing and re-analysis of data.1,2 This challenge has gained wide recognition leading to the development of standardised data and metadata storage formats such as the ‘Open Microscopy Environment (OME)’.3
However, few standards exist for the reproducible analysis and re-use of image-derived properties themselves. Often non-transparent, non-traceable data filtering and data point rejection procedures are applied to go from observations to conclusions. This inhibits data re-evaluation leading to low reproducibility of the data analysis part of a study. Robust storage formats that allow for simple, transparent and convenient analysis, classification, and storage of image-derived data are needed to solve this reproducibility crisis.
The Solution: Mars
Recognising the reproducibility crisis in the field of single-molecule imaging, we developed Molecule Archive Suite (Mars): an open-source platform for storage of image-derived properties of biomolecules. Mars provides a collection of ImageJ24 commands for processing images and image-derived properties based on a new Molecule Archive data storage architecture. Originally developed to overcome performance and reproducibility problems with very large datasets (10-100 GBs) derived from single-particle tracking 5, MoleculeArchives store data as collections of individual biomolecule properties and image metadata records (figure 1). During creation, all records are assigned universally unique IDs (UUIDs) which serve as the primary keys for retrieval and storage. This framework allows for seamless merging of datasets as well as multithreaded processing of virtually stored archives.
A simple, yet powerful user interface built on this architecture (Mars Rover, figure2) allows for a fully trackable, precise, and fast data analysis process based on arbitrary properties. These properties can be calculated using Mars commands (f.e. kinetic change points, variance, intensity), using the advanced plotting options within Mars Rover (f.e. distance travelled, speed), or using customised scripting. UUIDs ensure the history of each record remains traceable through long and complex analysis pipelines involving numerous data filtering and merging steps, and at the same time an automatically generated analysis log keeps track of all executed calculations and filter criteria applied. The implementation of Mars in single-molecule studies allows for an easy, fast and above all reproducible analysis of image-derived properties and will lead to clear and well-substantiated conclusions from single-molecule observations.
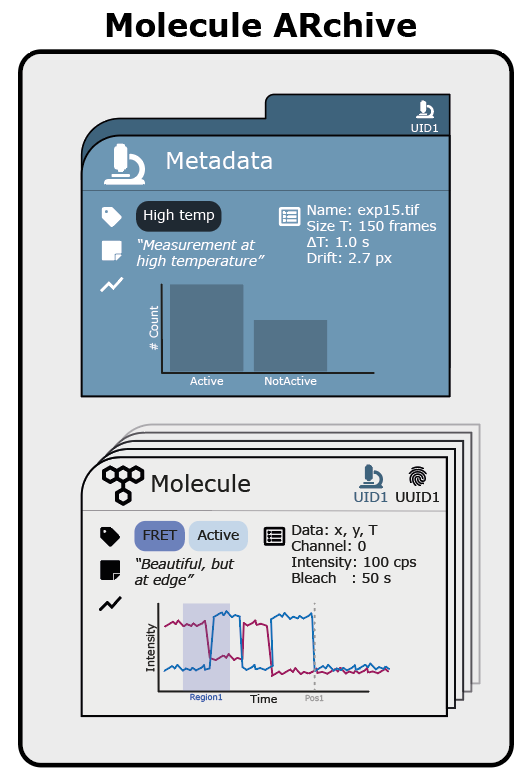
Figure 1: Schematic overview of the Molecule Archive architecture comprising 1. individual molecule records (grey cards) containing information such as molecule UUID, metadata UID, tracking coordinates, tags, notes, advanced plots and properties like intensity and bleaching time & 2. metadata records (blue card) containing experiment specific information like experiment dimensionality, frame rate, drift, microscope properties, tags, notes, advanced plots etc.
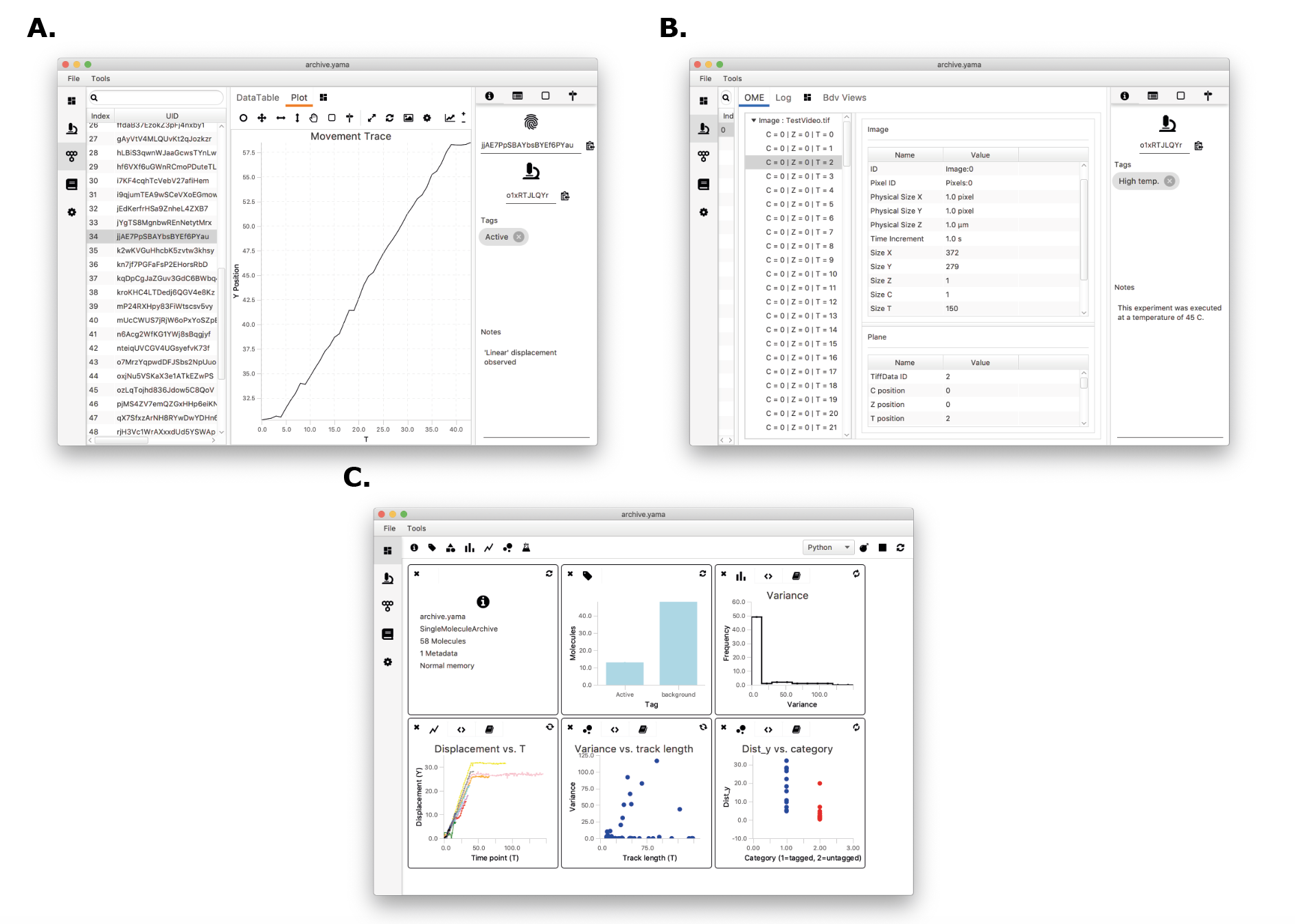
Figure 2: Screenshot of the Mars Rover user interface showing A. an individual molecule record showing tracking results, the molecule UUID, metadata UID, tags, and notes; B. a metadata record showing the metadata UID, microscope properties and experiment dimensionality; and C. the Rover Dashboard with scriptable widgets that can be customised fully according to the needs of the user.
Start using Mars
Please visit the install section to get Mars running on your machine and start analysing your data in a clear, reproducible manner. Visit the tutorials section for a head start and go through the extensive documentation pages for a more detailed description of each Mars command.
References
1 Marqués, G., Pengo, T., Sanders, M.A. (2020), “Science Forum: Imaging methods are vastly underreported in biomedical research”, eLife 9: e55133
2 Cooper, N., Hsing, P-J. (2017), “Guides to Better Science: Reproducible Code”, Britisch Ecological Society
3 Goldberg, IG., Allan, C., Burel, J-M. et al. (2005), “The Open Microscopy Environment (OME) Data Model and XML file: open tools for informatics and quantitative analysis in biological imaging”, Genome Biology 6: R47
4 Rueden, CT.; Schindelin, J. & Hiner, MC. et al. (2017), “ImageJ2: ImageJ for the next generation of scientific image data”, BMC Bioinformatics 18: 529, PMID 29187165
5 Agarwal et al. (2020), in revision.