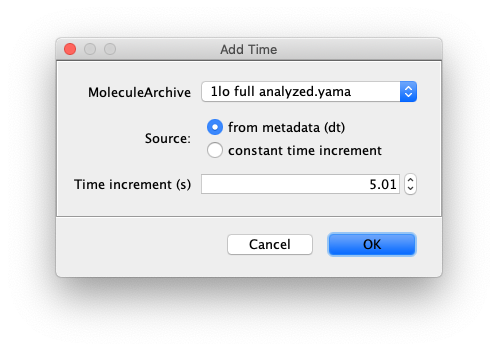
Add Time
This commands adds a column to the molecule tables to convert time points (T) to real time values in seconds. To do so the user can either select the dt value specified in the metadata or define a time increment themselves. This is a mapping procedure that is not disturbed by missing data points.
Inputs
- MoleculeArchive - MoleculeArchive input for the time conversion.
- Source - Select either the ‘dt’ information specified in the metadata are the option ‘Time increment’ in case the time increment will be specified below.
- Time increment (s) - Input value for the time increment if selected above.
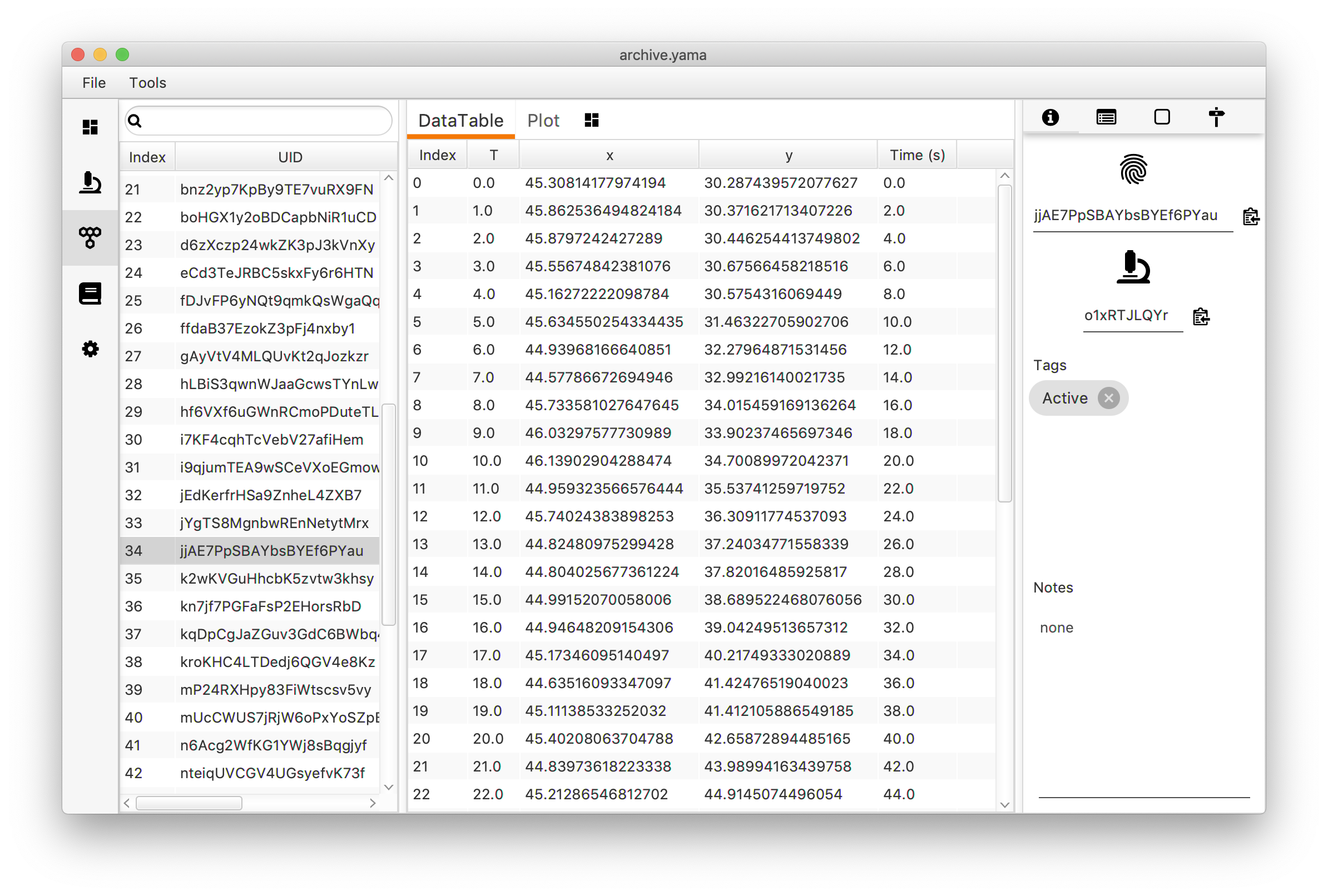
Output
- MoleculeArchive - Modified MoleculeArchive that was provided as input, the column ‘Time (s)’ is added to each molecule table.
How to run this Command from a groovy script
#@ MoleculeArchive archive
#@ ImageJ ij
import de.mpg.biochem.mars.molecule.*;
//Make an instance of the Command you want to run.
final AddTimeCommand addTime = new AddTimeCommand();
//Populates @Parameters Services etc. using the current context
//which we get from the ImageJ input.
addTime.setContext(ij.getContext());
//Set all the input parameters
addTime.setArchive(archive);
addTime.setSource("dt");
addTime.setTimeIncrement(2);
//Run the Command
addTime.run();
//Retrieve output from the command
//In this case, the archive is just modified
//so no output is needed and the line below is not needed
//archive = regionDifference.getArchive();