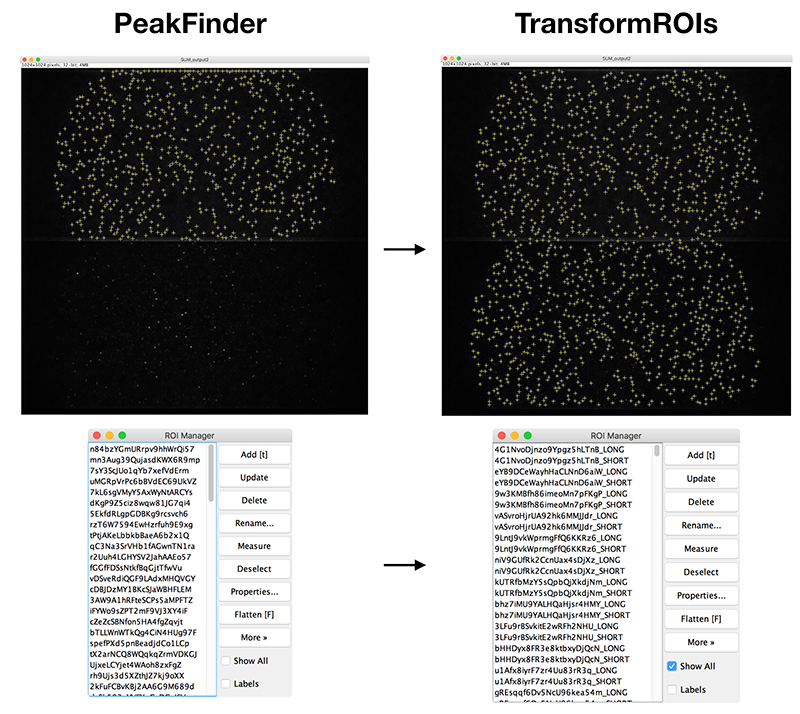
Transform ROIs
This command is used to transform ROIs from one region of an image to another for processing images collected using a multiview setup where different wavelengths are separated onto different regions of a camera sensor. A list of point or circle ROIs created using the Peak Finder with output option ‘Add to ROI Manager’ must be provided in the ROI Manager as setup prior to running this command. The ROIs are transformed using the Affine2D transformation parameters provided in the dialog. These can be generated using the Descriptor-based registration plugin with a sample containing beads that are fluorescent for all wavelengths. The Transform ROIs command duplicates the ROIs, transforms them, and updates all ROI names to include a region suffix. This command can be performed multiple times if the camera sensor has more than two regions to support triple and quadview setups. After running this command, the ROIs in the ROI Manager are the required input for the Molecule Integrator (multiview).
The colocalize tab allows for filtering of ROIs in one channel based on the fluorescent intensity in the other channel. This can be used, for example, to create a set of ROIs for molecules bound to DNA by detecting fluorescence in the DNA channel and filtering the original ROIs by this detected fluorescence.
Input
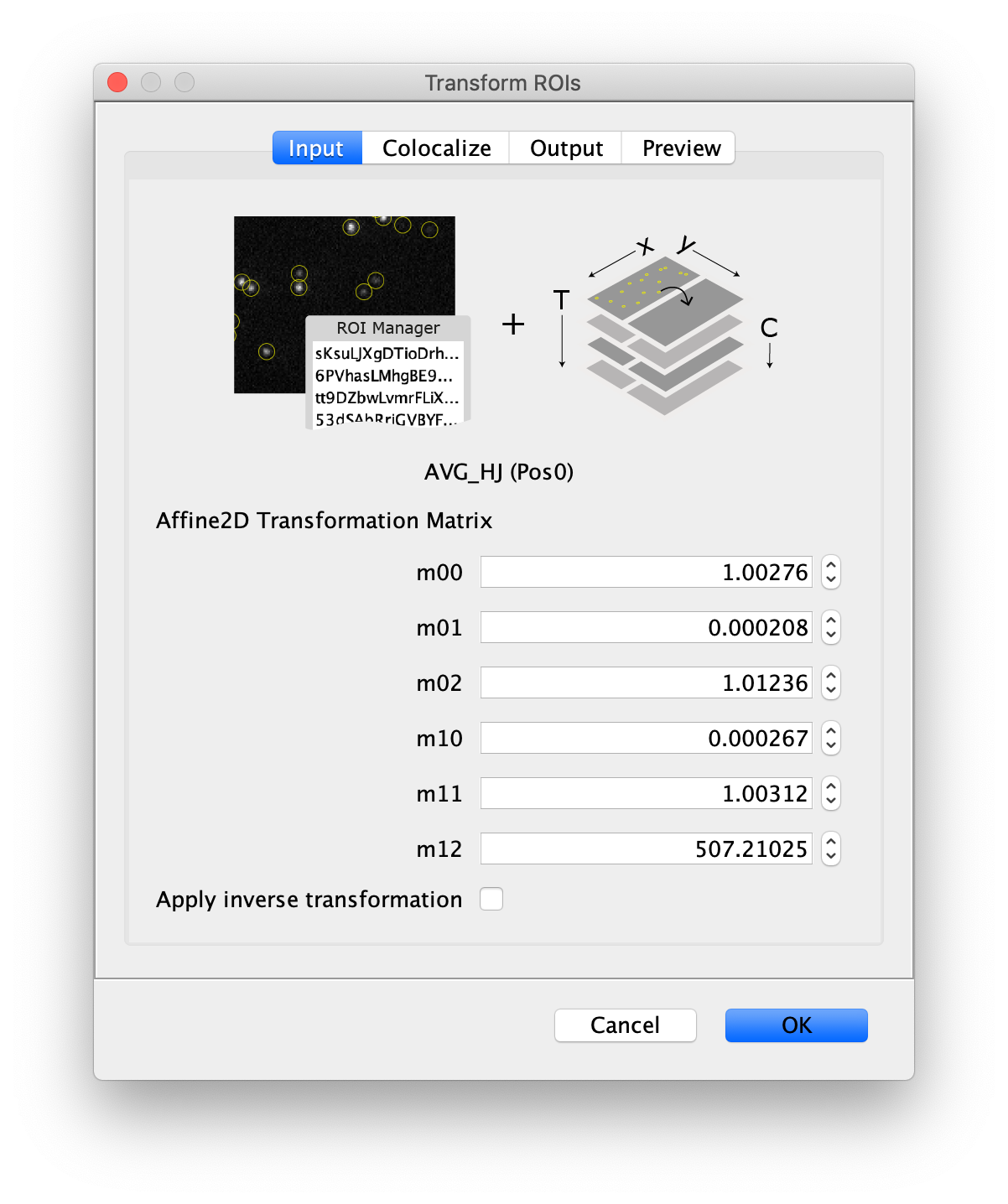
- Image - The selected image will by analyzed using the peaks in the ROI Manger. The name of the image is displayed below the image stack in the dialog window.
- Affine2D Transformation Matrix - The matrix values for the Affine2D transform generated using the Descriptor-based registration plugin.
- m00 - m00 matrix value.
- m01 - m01 matrix value.
- m02 - m02 matrix value.
- m10 - m10 matrix value.
- m11 - m11 matrix value.
- m12 - m12 matrix value.
- Apply inverse transformation - When checked the inverse transform will be applied to the ROIs.
Colocalize
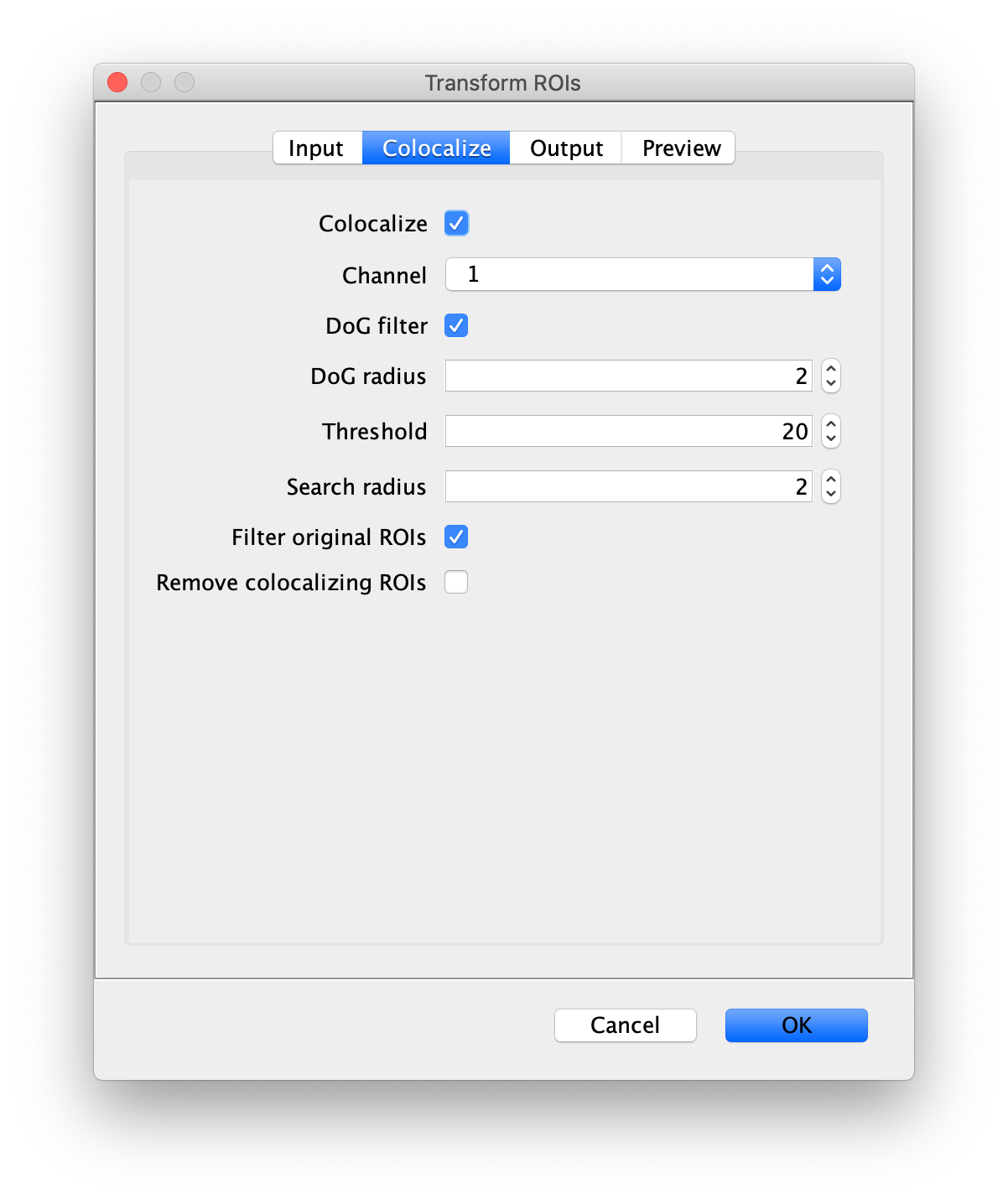
- Colocalize - If checked the transformed peak positions will be checked for peaks and filtered to remove transformations to locations that do not have peaks in the second channel.
- Channel - Select the channel that needs to be transformed in the case of a multi-channel video.
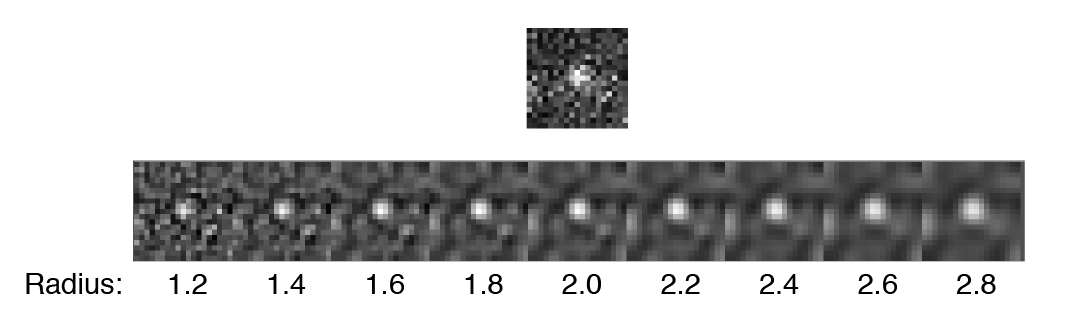
- DoG filter - If checked the image will be processed with a Difference of Gaussian (DoG) filter before peak finding. Using an appropriately chosen radius this filter enhances real peaks with signal spread among several pixels and suppresses salt and pepper noise as demonstrated in this systematic study. If unchecked the raw image will be used for peak finding.
- DoG radius - The radius used for DoG filtering. The value chosen should reflect the size of the desired peaks. Decimal numbers are permitted.
- Threshold - This is threshold in pixel value used for peak detection. Pixels with values above this threshold are considered peaks and pixels below this threshold are considered background. For everyday peak detection, the DoG filter should be used in which case this threshold using on the DoG filtered image. In this case, we have found that values between 40 and 60 provide optimal detection for typical single-molecule observations. If the DoG Filter is turned off this threshold reflects raw pixel values in the input image.
- Search radius - The radius of pixels to include for peak detection in the transformed peak. If the alignment is not perfect or the signal is more distributed in the transformed position, this setting will ensure there are fewer false negatives.
- Filter Original ROIs - If checked the peaks detected at the transformed locations will be used to filter the original ROIs in the starting channel. This is helpful to remove all proteins that do not colocalize with DNA for example. The effect will not be shown in the preview, it will only be show in the resulting output ROIs.
- Remove colocalizing ROIs - Remove ROIs in case the positions colocalize. This can be of use to filter out all peaks observed in both parts of the dualview resulting in a set of peaks that do not overlap and hence only have one of the two fluorophores. Note: This feature can only be enabled when ‘Filter Original ROIs’ is enabled.
Output
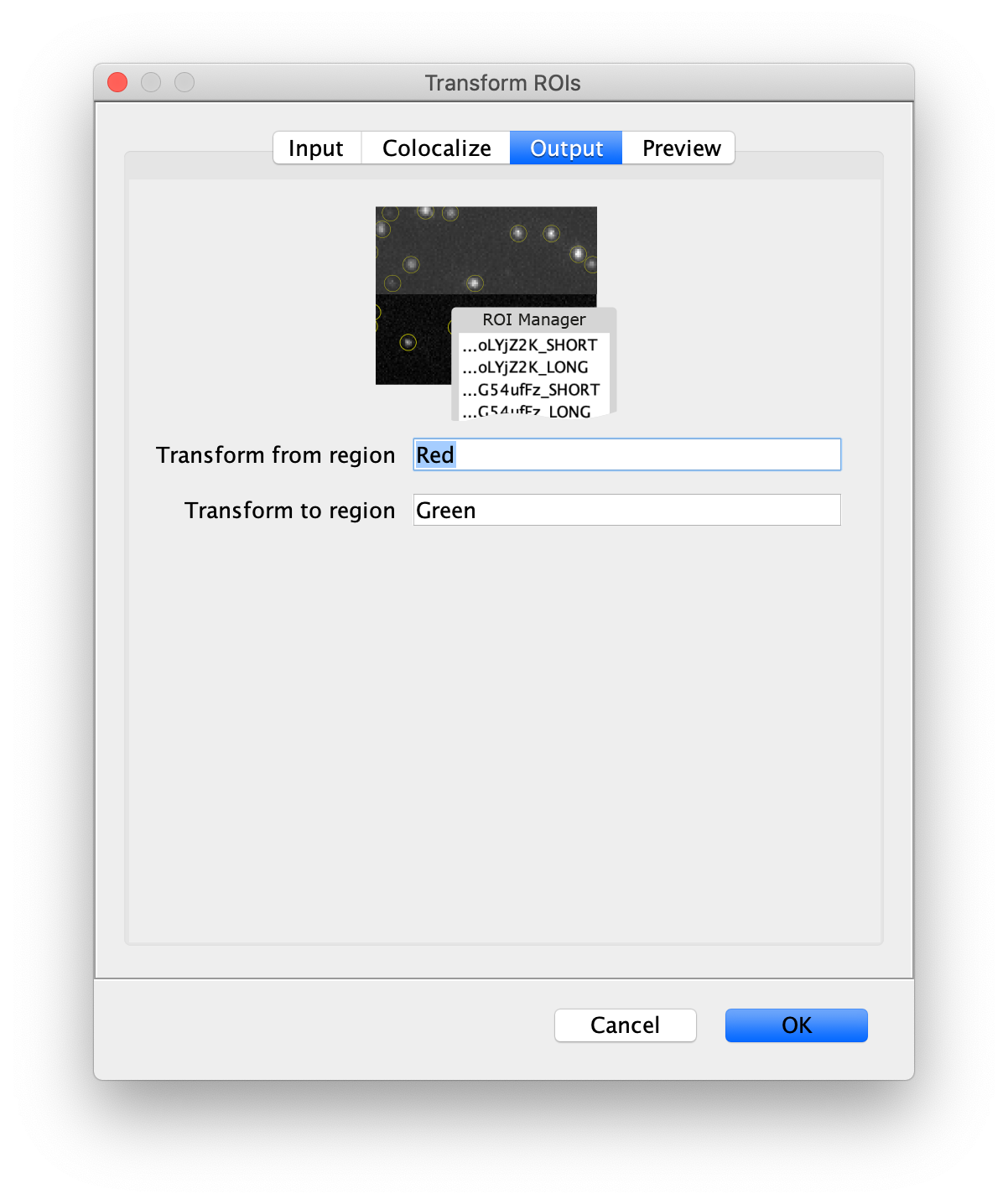
- Transform from region - The name of the region that ROIs are being transform from. If the ROIs in the ROI Manager do not have region suffixes, this region name will be added as a suffix to original ROI names. If the ROIs in the ROI Manager have region suffixes already, a choicebox is provided to select the region that should be transformed. In this way the command can be run several times in sequence to support triple and quadview setups.
- Transform to region - The name of the region where ROIs are being transformed. This is added as a suffix to the transformed ROIs.
Preview
- Preview - If checked the resulting transformed peaks will show up on the image based on the settings. Note: The results of Filtering the original ROIs will not show up in preview but only in the final output.
- T - Select the frame (T) in case of a multi-frame video.
- Preview timeout (s) - Maximum time that is allowed to pass before an (unsuccessful) preview attempt will be terminated.
Output
- ROIs in the ROI Manager - This command manipulates ROIs in the ROI Manager by duplicating them, transforming them, and adding the region suffix names provided.
How to run this Command from a groovy script
Simple example script transforming ROIs for two regions. The Affine2D transformation coordinates were generated based on sample data using a 1024 by 1024 camera with a dualview splitting the emission to the top and bottom of the image. As seen in the example image at the top of this page. Can you generate the Affine2D transformation coordinates for your image using the Descriptor-based registration plugin.
This script requires an instance of the ROI Manager so it will likely not work in headless mode.
#@ ImageJ ij
#@ RoiManager roiManager
import de.mpg.biochem.mars.roi.commands.*
//Make an instance of the Command you want to run
final TransformROIsCommand transformROIs = new TransformROIsCommand()
//Populates @Parameters Services etc. in the command using the current context
//which we get from the ImageJ input
transformROIs.setContext(ij.getContext())
transformROIs.setRoiManager(roiManager)
transformROIs.setM00(1.00276)
transformROIs.setM01(0.000208)
transformROIs.setM02(1.71236)
transformROIs.setM10(0.000267)
transformROIs.setM11(1.00312)
transformROIs.setM12(506.91025)
transformROIs.setTransformFromRegion("Red")
transformROIs.setTransformToRegion("Green")
//In this script we transform all ROIs without checking for colocalization
transformROIs.setColocalize(false)
//Run the Command
transformROIs.run()
This example script is similar to the one above but with an added filter for peaks based on colocalization.
#@ Dataset dataset
#@ ImageJ ij
#@ RoiManager roiManager
import de.mpg.biochem.mars.roi.commands.*
//Make an instance of the Command you want to run
final TransformROIsCommand transformROIs = new TransformROIsCommand()
//Populates @Parameters Services etc. in the command using the current context
//which we get from the ImageJ input
transformROIs.setContext(ij.getContext())
transformROIs.setRoiManager(roiManager)
transformROIs.setDataset(dataset)
transformROIs.setM00(1.00276)
transformROIs.setM01(0.000208)
transformROIs.setM02(1.71236)
transformROIs.setM10(0.000267)
transformROIs.setM11(1.00312)
transformROIs.setM12(506.91025)
transformROIs.setTransformFromRegion("Red")
transformROIs.setTransformToRegion("Green")
transformROIs.setChannel(1)
transformROIs.setT(0)
transformROIs.setUseDogFilter(true)
transformROIs.setDogFilterRadius(2)
transformROIs.setThreshold(50)
transformROIs.setFilterOriginalRois(true)
transformROIs.setFilterColocalizingRois(false)
transformROIs.setColocalizeSearchRadius(2)
//Run the Command
transformROIs.run()