Build archive from table
This command builds a Molecule Archive using a MarsTable. The Table>Open Table command can open csv formatted tables or MarsTables (json or yamt format). This command, together with the Open Table command, provides the possibility of building a Molecule Archive using csv data imported from another platform.
Inputs
- Table - MarsTable opened using the Table>Open Table command that accepts csv, json, and yamt.
- Molecule id column - Table column with unique ids for sets of rows representing each molecule record. The id column could contain integer index values (like in the image below), double values, or id strings. Select “None” if the table only contains data for one molecule.
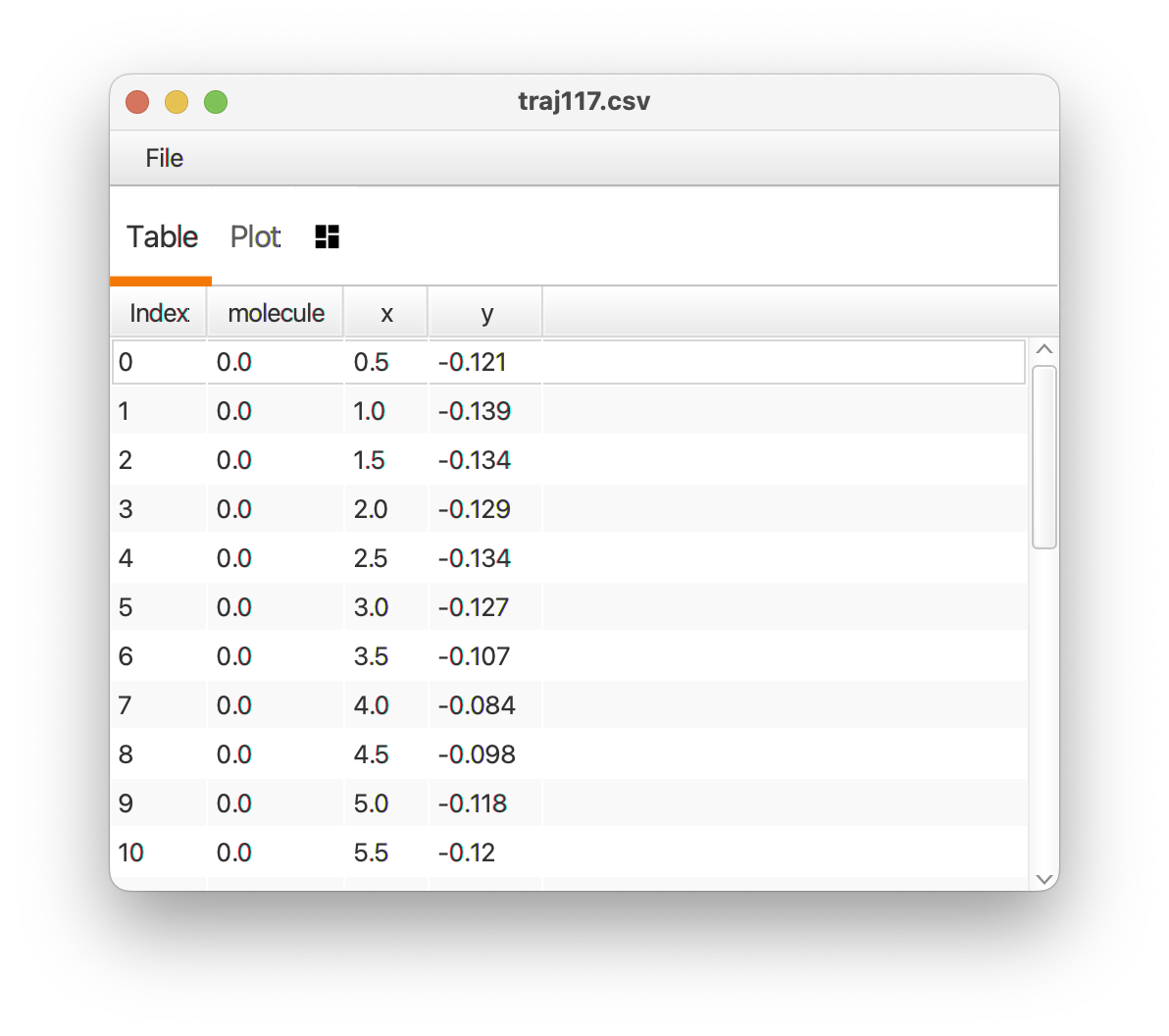
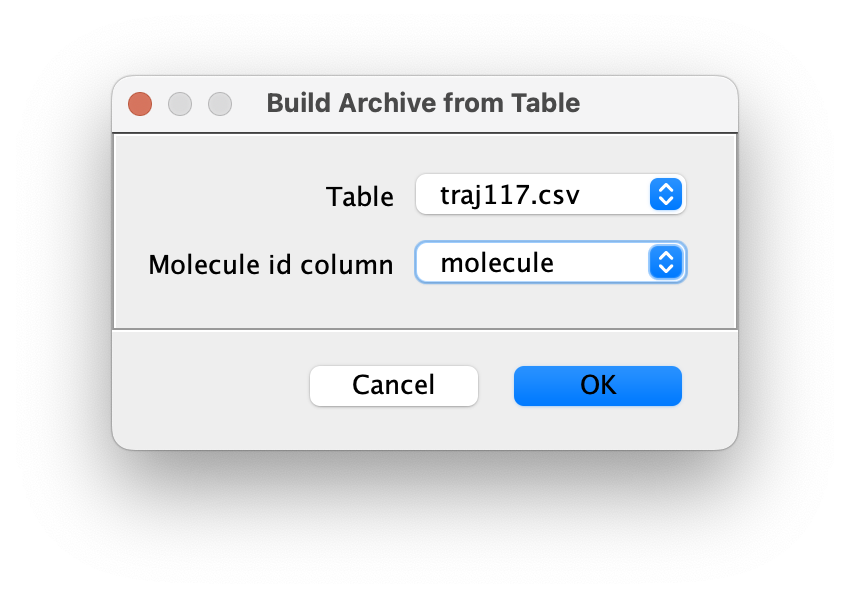
Output
- A MoleculeArchive with molecule records storing the MarsTable data. If a molecule id column is provided, rows with unique ids are placed in different molecule records. If no molecule id column is provided a single molecule record is created with all table data.
How to run this Command from a groovy script
#@ MarsTable table
#@ ImageJ ij
#@output MoleculeArchive archive
import de.mpg.biochem.mars.molecule.*
import de.mpg.biochem.mars.table.*
//Make an instance of the Command you want to run...
final BuildArchiveFromTableCommand archiveFromTable = new BuildArchiveFromTableCommand()
//Populates @Parameters Services etc.. using the current context
//which we get from the ImageJ input...
archiveFromTable.setContext(ij.getContext())
//Set all the input parameters
archiveFromTable.setTable(table)
archiveFromTable.setMoleculeIdColumn("molecule")
//Run the Command
archiveFromTable.run()
//Retrieve output from the command
archive = archiveFromTable.getArchive()
Alternatively, you can just use the MoleculeArchive constructer for creating an archive from a MarsTable:
#@ MarsTable table
#@ MoleculeArchiveService moleculeArchiveService
#@output MoleculeArchive archive
import de.mpg.biochem.mars.molecule.*
import de.mpg.biochem.mars.table.*
//Assumes all table data should go into a single molecule record
archive = new MoleculeArchive("archive.yama", table)
//OR
//The last input is the Molecule id column to use.
//archive = new MoleculeArchive("archive.yama", table, "molecule")