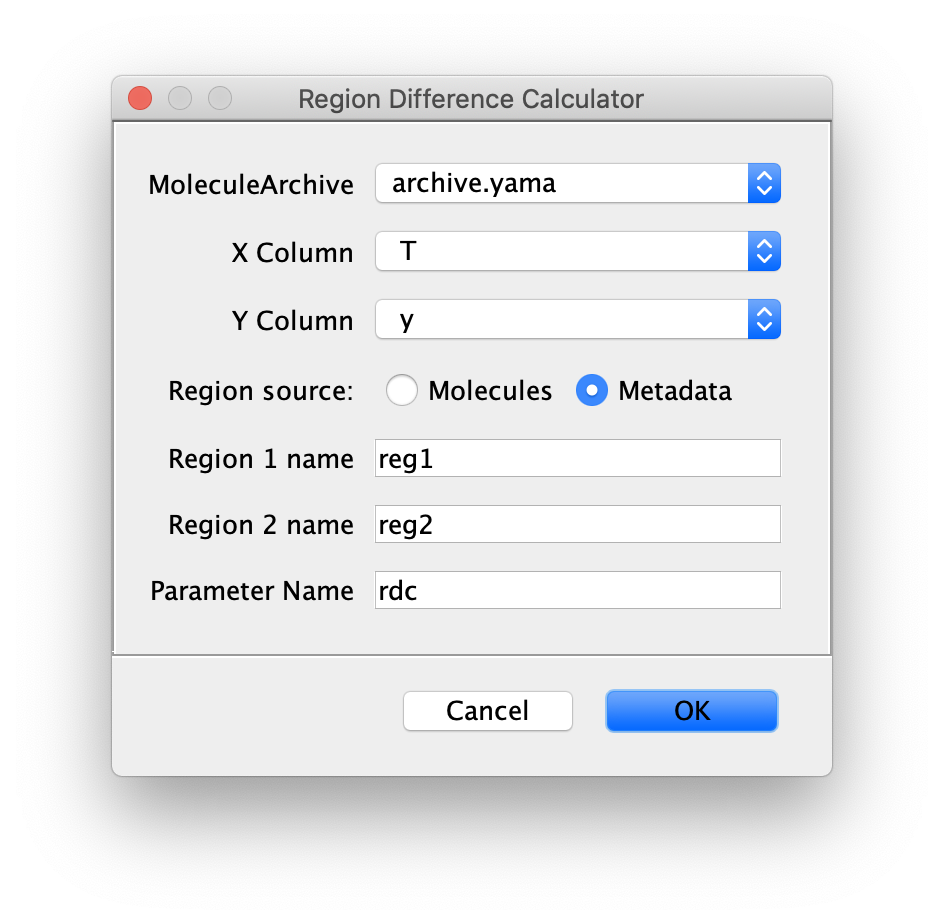
Region difference calculator
Inputs
- MoleculeArchive - MoleculeArchive input to use for the difference calculation. This command will calculate the difference between the regions specified for all molecules in the archive and add the values for each molecule as the parameter specified below.
- X Column - X column for the purposes of specifying the reversal regions.
- Y Column - Y column used for calculating the difference.
- Region source - Defines from which part of the Archive the region information is retrieved. Choose between regions defined globally in the metadata or locally defined regions in the molecules section.
- Region 1 name - Name of the first region.
- Region 2 name - Name of the second region.
- Parameter Name - Name of the Parameter added to each molecule entry in the archive with the calculated difference.
Output
- The MoleculeArchive provided as Input is modified.
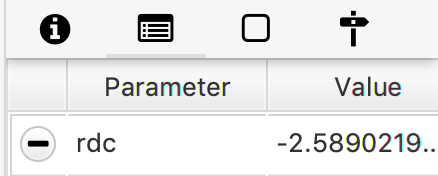
Running the command on an archive with this molecule results in the Parameter “rdc” with value -2.59 being added for this molecule.
How to run this Command from a groovy script
#@ MoleculeArchive archive
#@ ImageJ ij
import de.mpg.biochem.mars.molecule.*;
import de.mpg.biochem.mars.table.*;
//Make an instance of the Command you want to run...
final RegionDifferenceCalculatorCommand regionDifference = new RegionDifferenceCalculatorCommand();
//Populates @Parameters Services etc.. using the current context
//which we get from the ImageJ input...
regionDifference.setContext(ij.getContext());
//Set all the input parameters
regionDifference.setArchive(archive);
regionDifference.setXcolumn("T");
regionDifference.setYcolumn("y");
regionDifference.setRegionSource("Metadata");
regionDifference.setRegionOne("reg1");
regionDifference.setRegionTwo("reg2");
regionDifference.setParameterName("rdc");
//Run the Command
regionDifference.run();
//Retrieve output from the command
//In this case, the archive is just modified
//so no output is needed and the line below is not needed
//archive = regionDifference.getArchive();