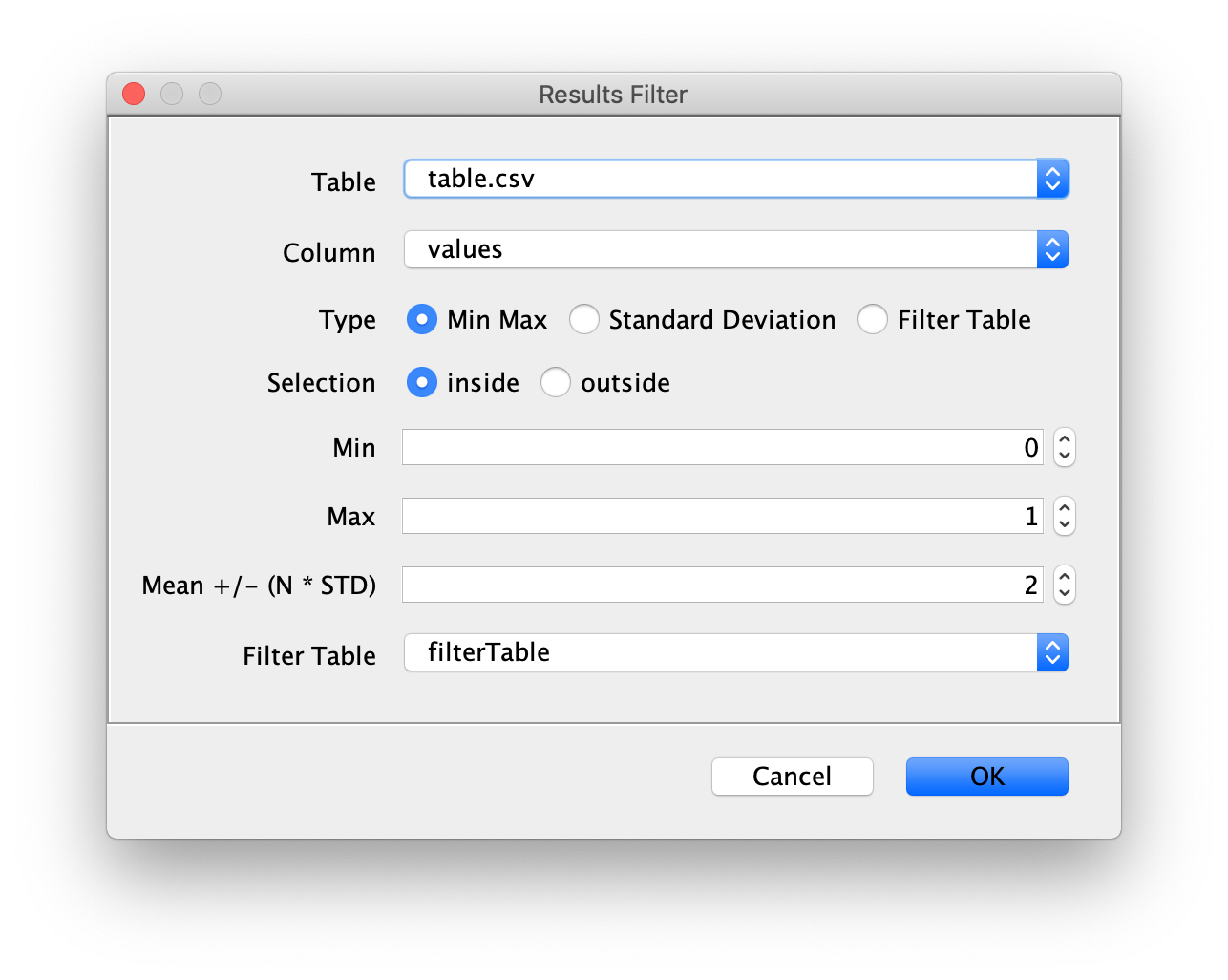
Filter
This command offers numerous ways to filter MarsTable rows based on their values. This includes simple Min/Max filtering, or filtering all values that lie outside some number of standard deviations from the mean, and filtering one table based on the values of another.
In all cases, the “inside” and “outside” options provide a choice between all values that lie within the specified region or all values that lie outside the specified region.
Inputs
- “Table” - The open MarsTable that you want to filter.
- “Column” - The column to use for filtering.
- “Type” - The type of filter to apply. If “Min Max” is selected, the table will be filtered based on the range given using the Min and Max values. The range is inclusive of the end points. If “Standard Deviation” is selected, the table will be filtered based on the range given by the “Mean +/- (N * STD)” value. This value is the N in the formula, Finally, if “Filter Table” is selected, the table will be filtered based on the values in the Filter Table provided. In this case, only matching values between “Column” in the Filter Table and “Column” in Table will be included or excluded.
- “Selection” - This selection of values to keep. If “inside” is selected then all values specified by the filter will be kept. If “outside” is selected then all values not specified by the filter will be kept.
- “Min” - The minimum value if using “Min Max” Filter.
- “Max” - The maximum value if using “Min Max” Filter.
- “Mean +/- (N * STD)” - The number of standard deviations from the mean to include in the range. The N value in the formula given. This value is used if “Standard deviation” is chosen as the filter type.
- “Filter Table” - A second table to use as a reference if type is set to “Filter Table.”
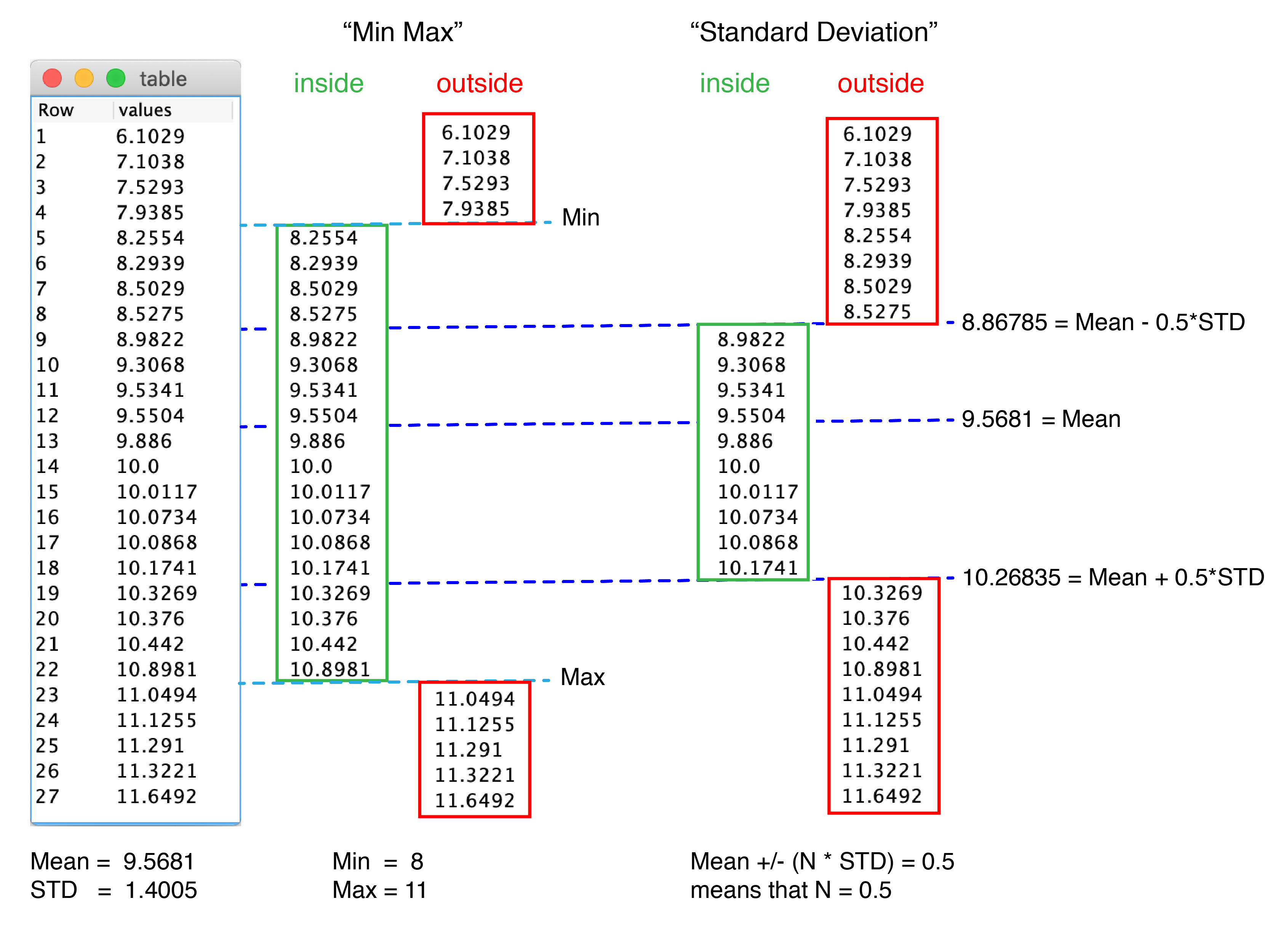
Example outputs depending on the options
Example for type “Filter Table” with “molecule” selected as the column.

Output
- A filtered MarsTable.
How to filter tables in a groovy script
#@ MarsTable table
#@ ImageJ ij
import de.mpg.biochem.mars.table.*
//Make an instance of the Command you want to run...
final ResultsTableFilterCommand resultsFilter = new ResultsTableFilterCommand()
//Populates @Parameters Services etc.. using the current context
//which we get from the ImageJ input...
resultsFilter.setContext(ij.getContext())
resultsFilter.setTable(table)
resultsFilter.setColumn("values")
//Options: "Min Max" , "Standard Deviation" , "Filter Table"
resultsFilter.setType("Min Max")
//Options: "inside" , "outside"
resultsFilter.setSelection("inside")
//If "Min Max"
resultsFilter.setMin(0)
resultsFilter.setMax(300)
//If "Standard Deviation" then uncomment
//resultsFilter.setNSTD((double)2)
//If "Filter Table", then uncomment and provide a filterTable
//resultsFilter.setFilterTable(filterTable)
//Run the Command
resultsFilter.run()