Build DNA archive
This command builds a DNA archive from a SingleMoleculeArchive and a list of DNA ROIs in the ROI manager. It uses the location of DNA molecules that were found with the DNA finder tool and searches for molecules in the SingleMoleculeArchive that overlap with (parts of) this location. This gives the user a great option to investigate protein movement along an immobilized DNA template. An example that extensively uses this tool can be found here.
Input
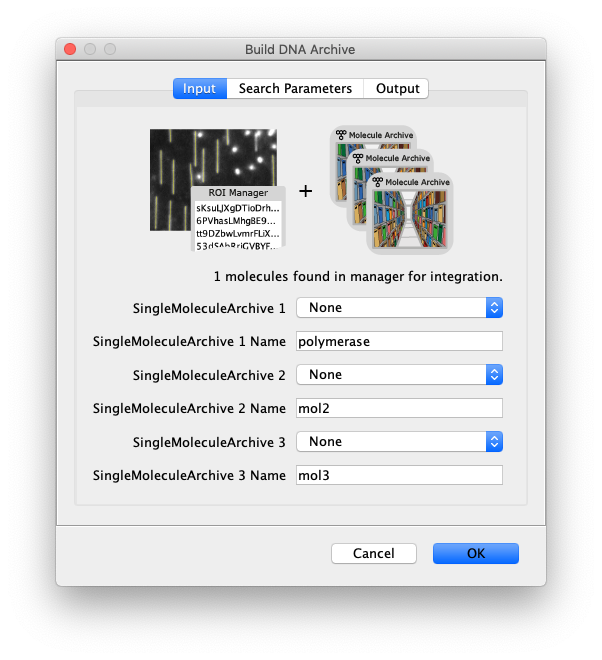
- SingleMoleculeArchive 1 - Select the first SingleMoleculeArchive to be matched to these DNA molecules.
- SingleMoleculeArchive 1 Name - Enter the name of the MoleculeArchive (f.e. the name of the measured protein that is tracked in this archive).
- Merge - Add a second SingleMoleculeArchive to the DNA archive. Supply the archive and name below.
- SingleMoleculeArchive 2 - Select the second SingleMoleculeArchive to be matched to these DNA molecules.
- SingleMoleculeArchive 2 Name - Enter the name of the MoleculeArchive (f.e. the name of the measured protein that is tracked in this archive).
- Merge - Add a third SingleMoleculeArchive to the DNA archive. Supply the archive and name below.
- SingleMoleculeArchive 3 - Select the third SingleMoleculeArchive to be matched to these DNA molecules.
- SingleMoleculeArchive 3 Name - Enter the name of the MoleculeArchive (f.e. the name of the measured protein that is tracked in this archive).
Search Parameters
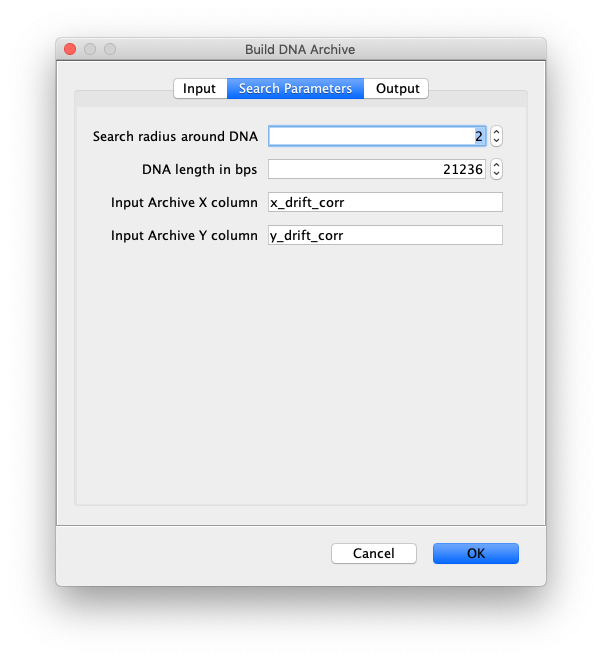
- Search radius around DNA - Defines the proximity limit in which the single molecule must be to be regarded overlapping with the DNA molecule.
- DNA length in bps - Enter the length of the imaged DNA in bps.
- Input Archive X column - Column of x-coordinates from the SingleMoleculeArchive to be regarded. This can be simply ‘x’ or for example ‘x_drift_corr’ in the case of a drift corrected archive.
- Input Archive Y column - Column of y-coordinates from the SingleMoleculeArchive to be regarded. This can be simply ‘y’ or for example ‘y_drift_corr’ in the case of a drift corrected archive.
Output
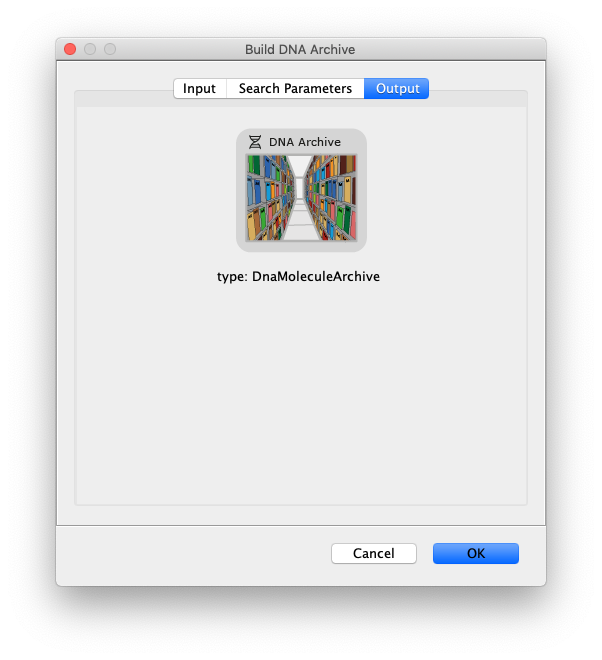
- DNA Archive - The output of this command is a DNA Archive that houses the location of the DNA molecules and the information of all SingleMolecule entries that are associated with these DNA molecules.
How to run this Command from a groovy script
#@ MoleculeArchive archive
#@ ImageJ ij
import de.mpg.biochem.mars.molecule.*;
//Make an instance of the Command you want to run.
final BuildDnaArchiveCommand dnaArchive = new BuildDnaArchiveCommand();
//Populates @Parameters Services etc. using the current context
//which we get from the ImageJ input.
addTime.setContext(ij.getContext());
//Set all the input parameters
dnaArchive.setDNASearchRadius(0)
dnaArchive.setDNALength(21236)
dnaArchive.setXColumn("x_drift_corr")
dnaArchive.setYColumn("y_drift_corr")
dnaArchive.setArchive1(archive1.yama)
dnaArchive.setArchive1Name("mol1")
dnaArchive.setArchive2(archive2.yama)
dnaArchive.setArchive2Name("mol2")
dnaArchive.setArchive3(archive3.yama)
dnaArchive.setArchive3Name("mol3")
//Run the Command
dnaArchive.run();