Variance Calculator
For an introductory tutorial on variance calculations in Mars the reader is referred to the tutorial.
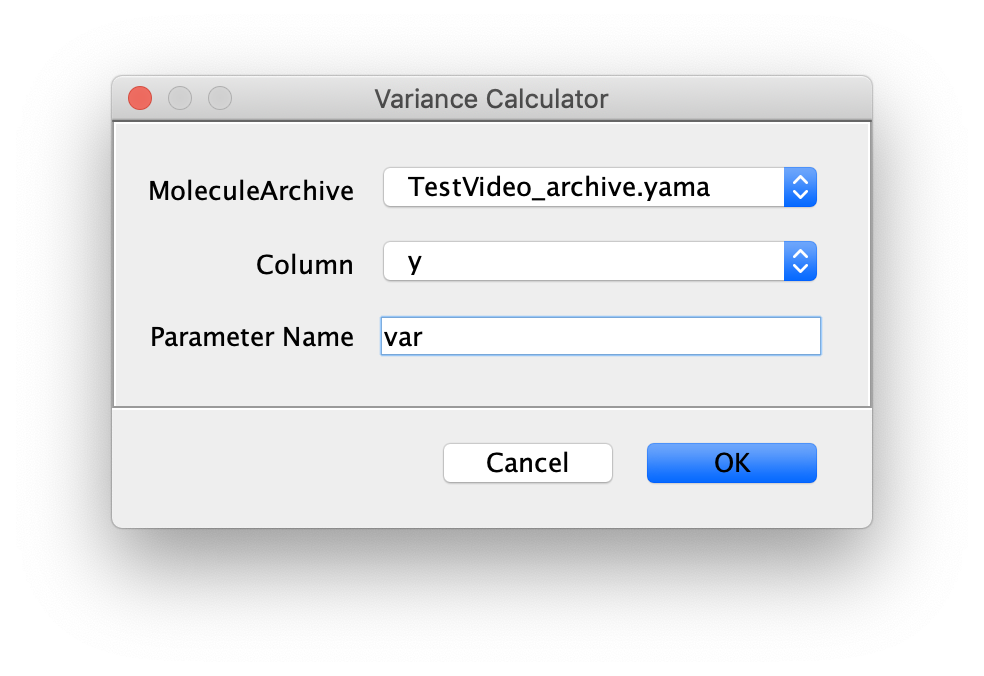
Inputs
- MoleculeArchive - MoleculeArchive to operate on.
- Column - Column of each molecule table to calculate the variance (var) for.
- Parameter Name - The name of the Parameter that will be added to each molecule record.
Output
- The calculated value is added as parameter to each molecule record in the MoleculeArchive.
How to run this Command from a groovy script
#@ MoleculeArchive archive
#@ ImageJ ij
import de.mpg.biochem.mars.molecule.*;
//Make an instance of the Command you want to run...
final VarianceCalculatorCommand varCalc = new VarianceCalculatorCommand();
//Populates @Parameters Services etc.. using the current context
//which we get from the ImageJ input...
varCalc.setContext(ij.getContext());
//Set all the input parameters
varCalc.setArchive(archive);
varCalc.setColumn("y");
varCalc.setParameterName("var");
//Run the Command
varCalc.run();