How to calculate an Affine2D matrix
level: intermediate, duration: 10-15 min
Introduction
To interpret data from a two channel dualvideo dataset obtained with single-molecule TIRF studies these two channels need to be aligned. The Descriptor-based registration (2d/3d) in Fiji provide robust tools for determination of coordinate transformations and can be used excellently to calculate these transformation coordinates. A video tutorial by the writer of this plugin explaining the basic features can be found on YouTube.
A dataset with bright fluorescent beads fluorescing in both channels is required for this calculation. This will then serve as a coordination framework that should remain equal for samples measured afterwards with the same laser and micro mirror alignment settings. Such an example dataset can be found on the repository.
How to run the command
First, open both images that should be aligned: image1 and image2 in this example. Next, locate the plugin in Fiji by using the search bar. Put in “registration” and select the “Descriptor-based registration (2d/3d)” plugin from the menu. Press run.
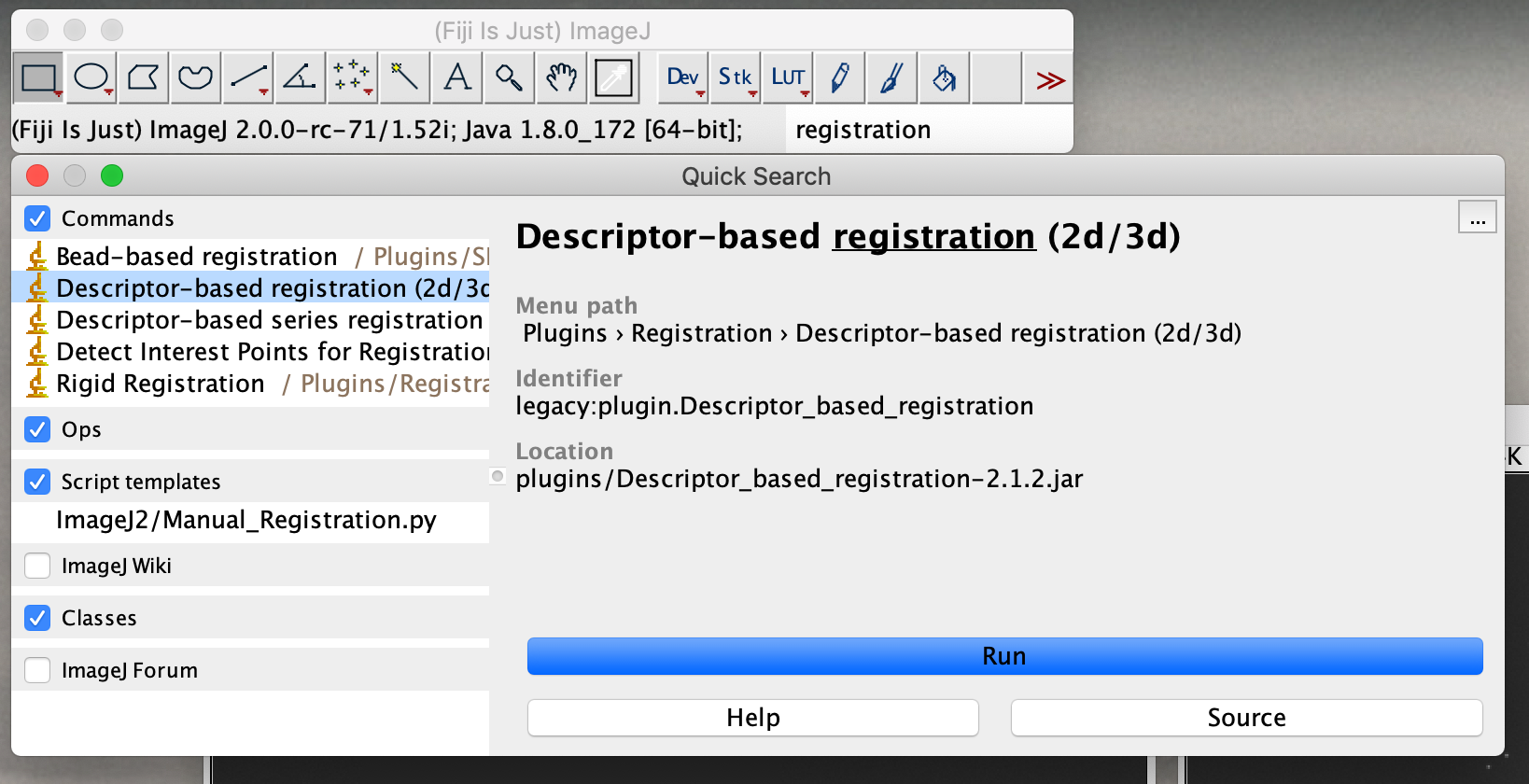
Select the two images of interest (in this example “image1.tif” and “image2.tif”) and press ok.
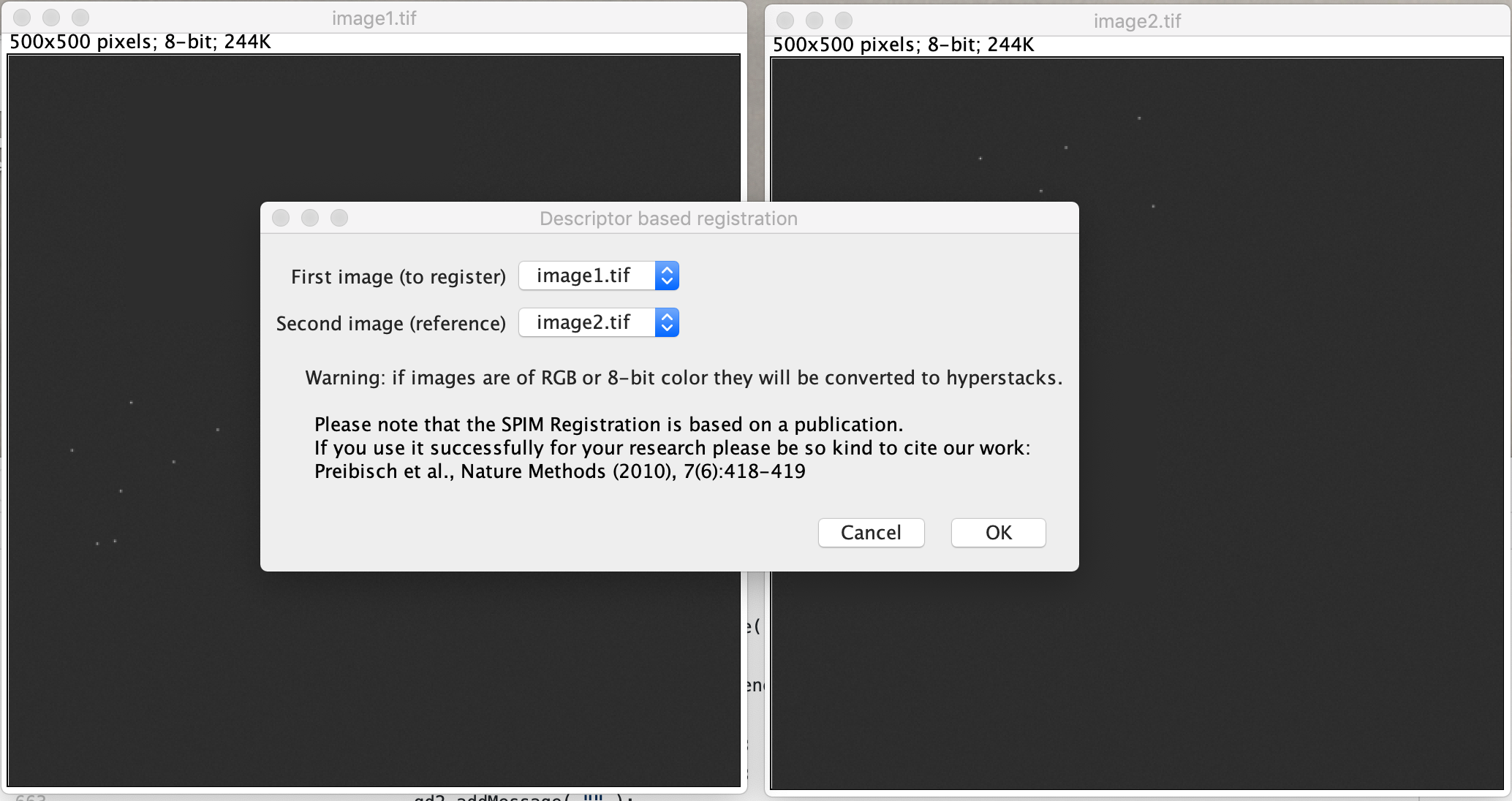
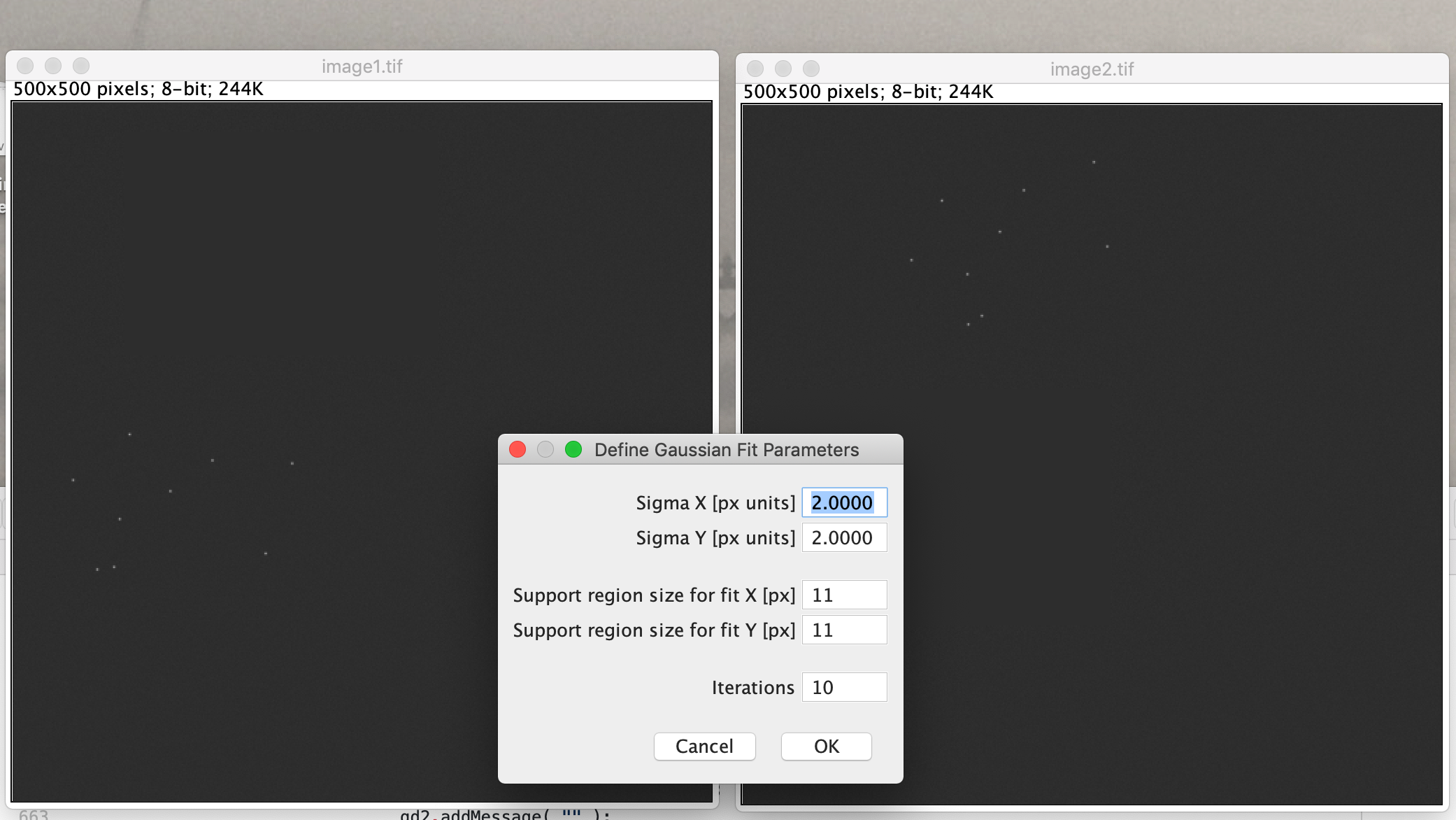
Select the settings as shown in the screenshot below. Select the ‘Gaussian mask localization fit’ for subpixel alignment, and next to that for our data we have been doing an 2D affine transform.
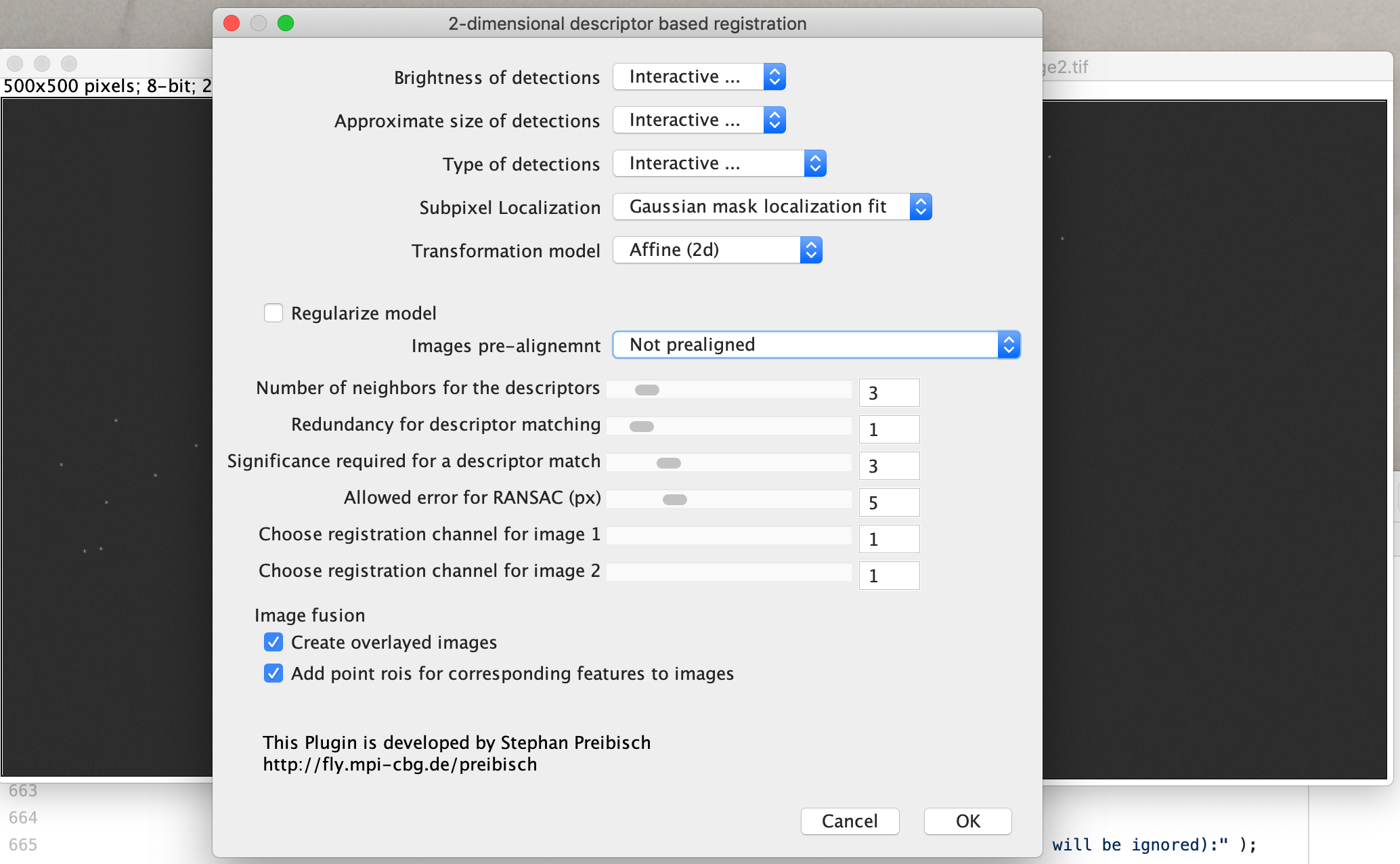
A yellow box will be displayed on image 1. Make sure that the fluorescent dots are within this box and show up as selected with the green indicators shown. Adjust sigma 1 and the threshold to find the optimum settings. Sigma 2 can only be applied when 3D data is interpreted.
Now look at the results shown in the fused image window. If everything went correctly the result for this particular example should look as follows.
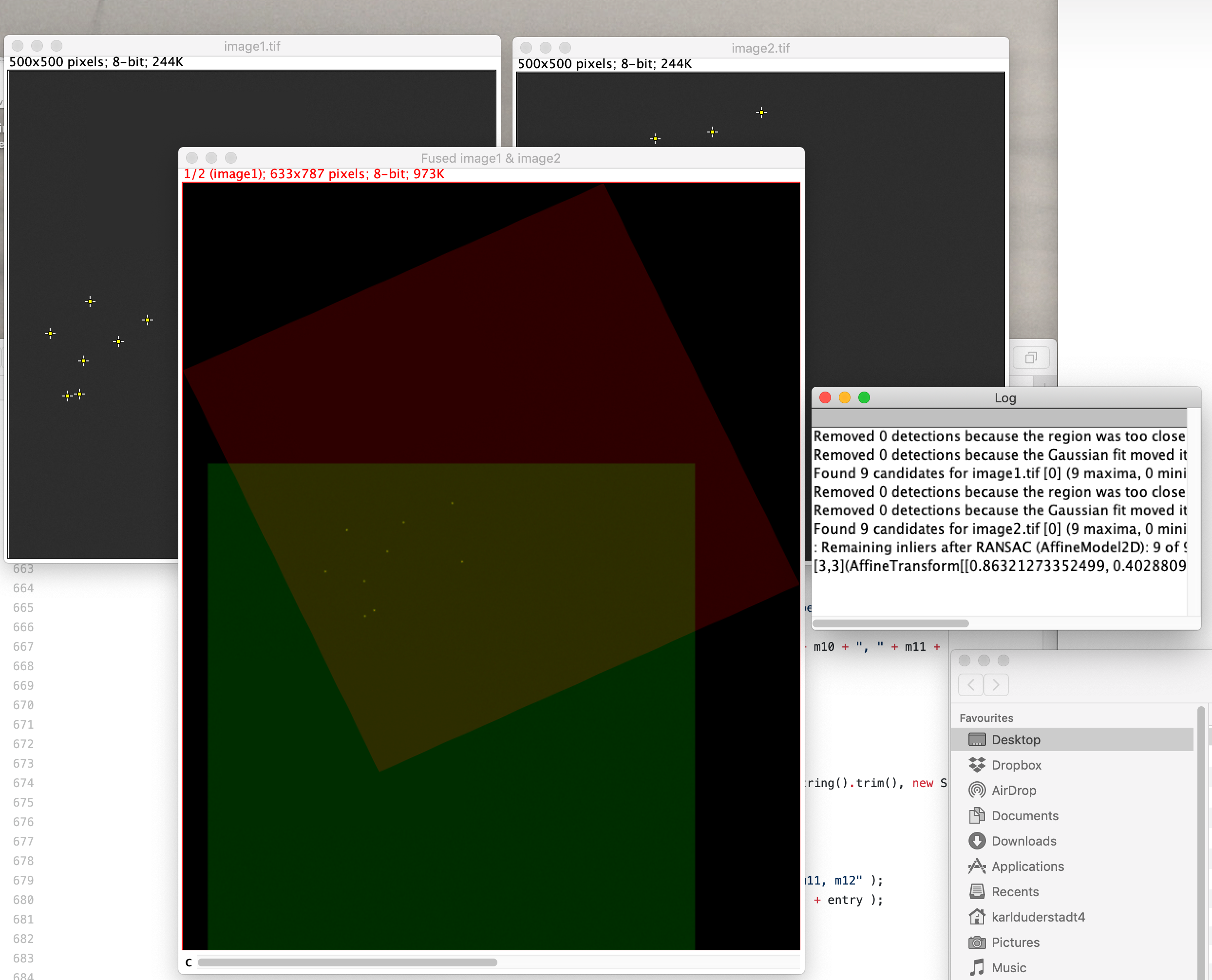
Besides a visual output, the console outputs the actual calculated coordinates. These are needed as an input in Mars Rover to use the BigDataViewer (see BigDataViewer tutorial) or when using the Transform ROIs command.