MarsTables tutorial
level: intermediate, duration: 10 min
This tutorial shows how to create and modify a MarsTable using the scripting feature in Fiji. MarsTables add the functionality of generating tables within the Fiji/ImageJ environment. For a general introduction on Groovy scripting or more advanced scripting the reader is referred to the scripting tutorials. A practical example of the use of a MarsTable is found in the Examples Gallery short examples, nr2.
1. How to create a MarsTable
This example shows how to create a MarsTable filled with dummy values. Open the script editor as explained in the introductory tutorial and run the following script.
#@output MarsTable table
import de.mpg.biochem.mars.table.*;
//Initialize a new ResultsTable with 3 columns and no rows
table = new MarsTable(3,0)
table.setColumnHeader(0, "column1")
table.setColumnHeader(1, "column2")
table.setColumnHeader(2, "column3")
for (int row=0;row<10;row++) {
//Since we initialised the table with no rows
//We have to increase the table size by one row
//before we can add more values
table.appendRow()
table.setValue("column1", row, row*0.25)
table.setValue("column2", row, row*0.25)
table.setValue("column3", row, row*0.25)
}
This generates the following MarsTable.
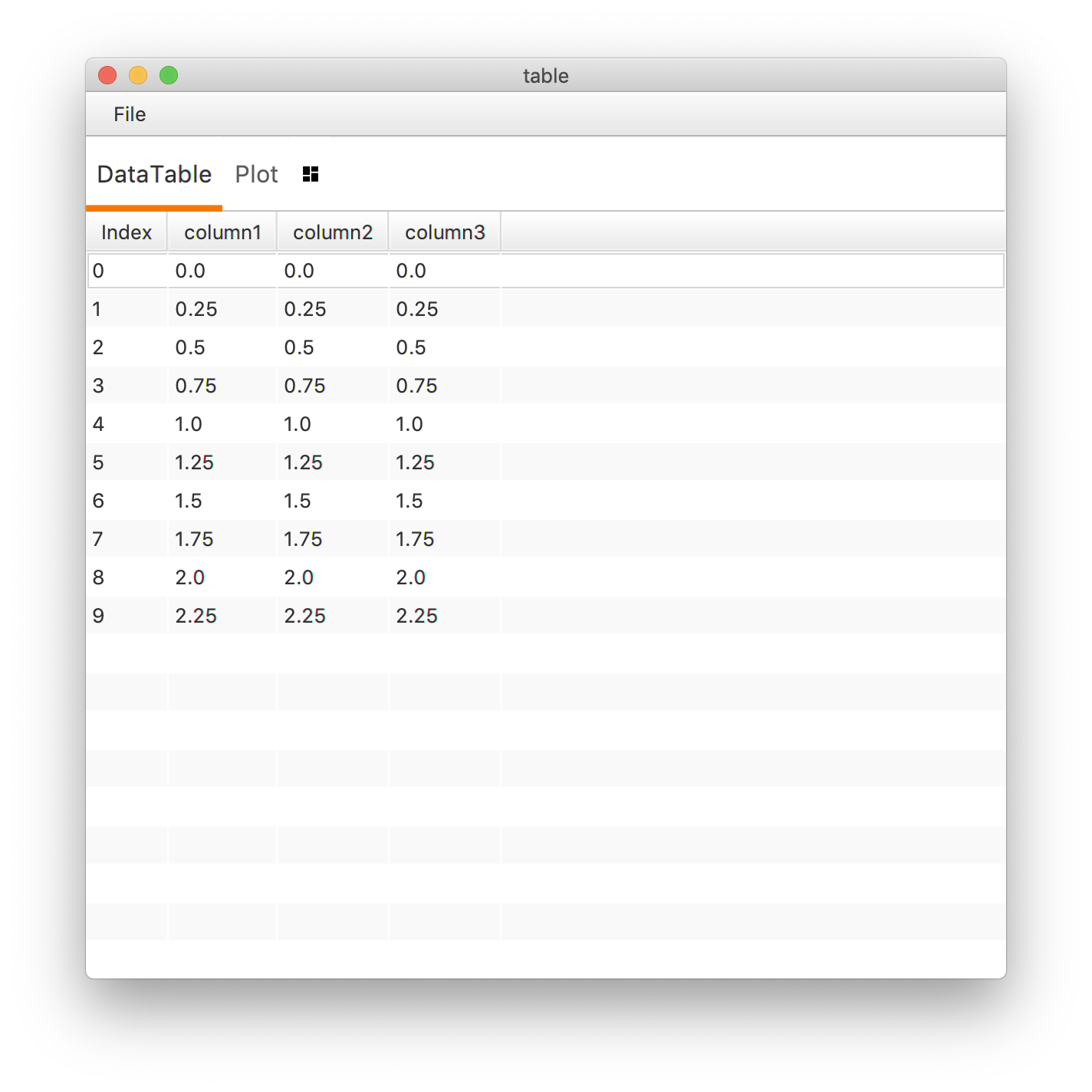
Alternatively, the same table can be generated by first building columns and then adding them to the table.
#@output MarsTable table
import de.mpg.biochem.mars.table.*
import org.scijava.table.*
//Initialize a new empty MarsTable
table = new MarsTable()
DoubleColumn col1 = new DoubleColumn("column1")
DoubleColumn col2 = new DoubleColumn("column2")
DoubleColumn col3 = new DoubleColumn("column3")
for (int row=0;row<10;row++) {
col1.add((double)row*0.25)
col2.add((double)row*0.25)
col3.add((double)row*0.25)
}
table.add(col1)
table.add(col2)
table.add(col3)
2. Adding columns to a MarsTable
After a MarsTable has been generated, it can be extended in the following two ways.
The first option is to create a new column, and then set the values in the column.
#@ MarsTable table
import de.mpg.biochem.mars.table.*
import org.scijava.table.*
double val = 1
table.appendColumn("molecule")
table.setValue("molecule", 0, val)
table.setValue("molecule", 1, val)
table.setValue("molecule", 2, val)
table.setValue("molecule", 3, val+1)
table.setValue("molecule", 4, val+1)
table.setValue("molecule", 5, val+1)
table.setValue("molecule", 6, val+2)
table.setValue("molecule", 7, val+2)
table.setValue("molecule", 8, val+2)
table.setValue("molecule", 9, val+2)
table.getWindow().update()
Alternatively, the following script can be used.
#@ MarsTable table
import de.mpg.biochem.mars.table.*
import org.scijava.table.*
DoubleColumn col = new DoubleColumn("molecule")
double val = 1
col.add(val)
col.add(val)
col.add(val)
col.add(val+1)
col.add(val+1)
col.add(val+1)
col.add(val+2)
col.add(val+2)
col.add(val+2)
col.add(val+2)
table.add(col)
table.getWindow().update()
In both cases the MarsTable is extended with the column named “molecule”.
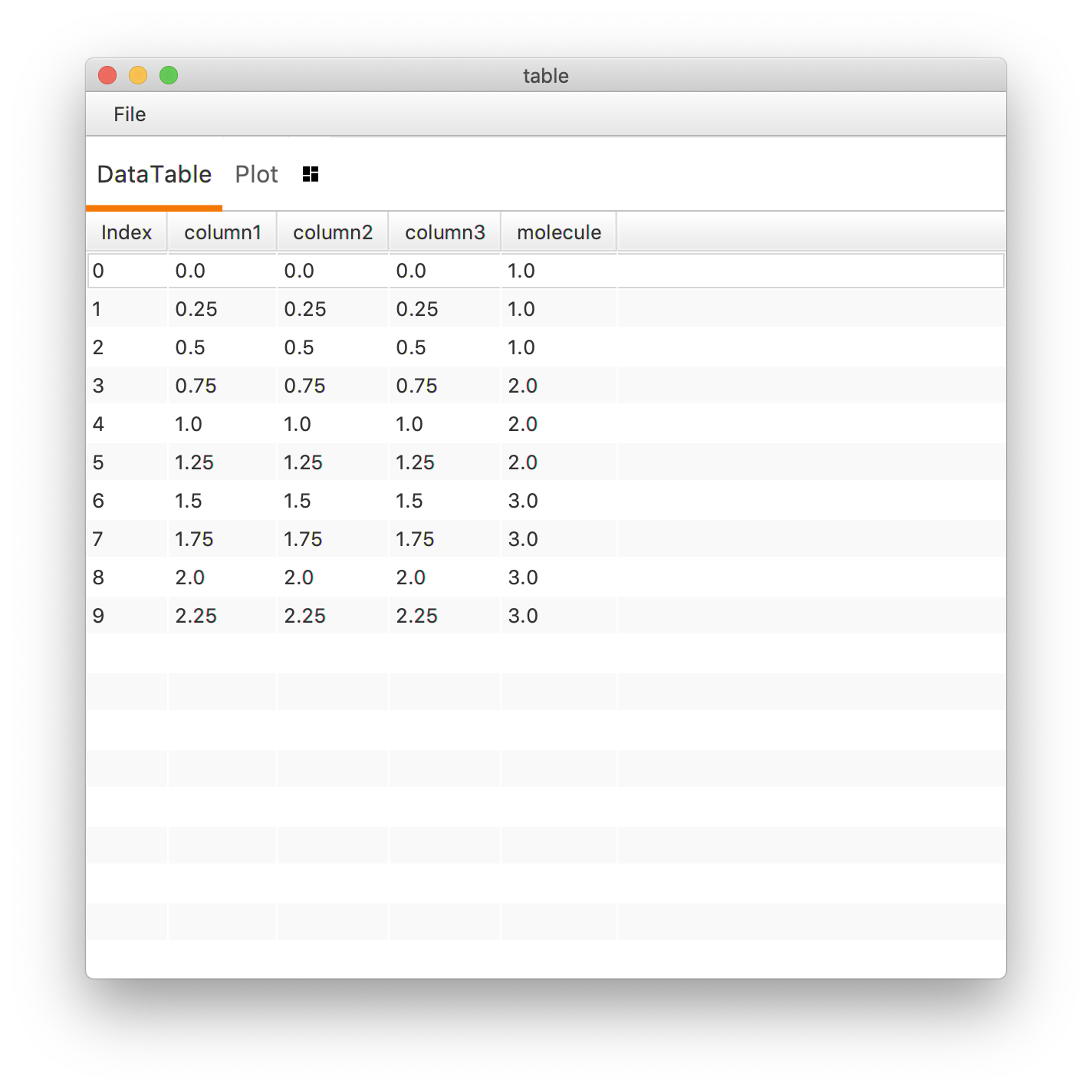
3. Remove a column from a MarsTable
To remove the newly added column “molecule” again use the following script.
#@ MarsTable table
import de.mpg.biochem.mars.table.*
import org.scijava.table.*
table.removeColumn("molecule")
table.getWindow().update()