Molecule Integrator
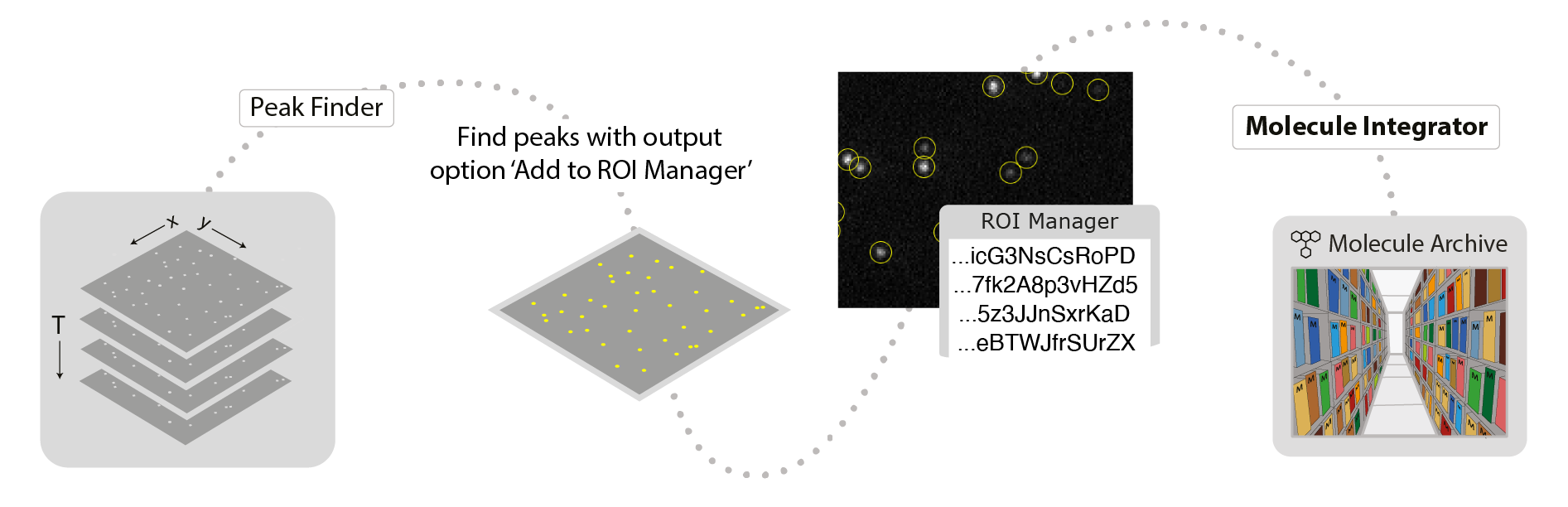
This command integrates the intensity of fluorescent peaks in images. A list of point or circle ROIs must be provided in the ROI Manager as setup prior to running this command. These are most easily generated using the PeakFinder with ‘Add to ROI Manager’ checked in the output tab. Otherwise, they can be manually added to the ROI Manager with unique names that will become the single molecule record UIDs. The active image will be the one integrated. The integration region for each peak is defined by inner and outer radii inputs. The inner radius defines the region of pixels to integrate and the outer radius defines the region used to determine the local background for correcting the intensity. Integration is performed for all peaks and all timepoints. The final output is a Single Molecule Archive with fluorescence intensity as a function of time point (T). This command assumes the molecule positions marked with ROIs remain constant for all time points. Therefore, only one entry is required for each molecule for each region in the ROI manager without the need to specify a time point. The single ROI entry is then integrated for all time points (T).
Use the Molecule Integrator (multiview) to integrate peaks from dual or multiview setups in which different emission wavelengths are separated onto different regions of the camera sensor. Using the Molecule Integrator (multiview) you can integrate multiple emission wavelengths and they are combined into single molecule records.
If you would like to integrate the fluorescence of molecules that are moving, you can use the PeakTracker. However, fluorescence will only be integrated for time points where peaks were detected, so no background will be integrated after bleaching. Integration maps can be provied to the Molecule Integrator (multiview) command in scripts with different positions for each time point for a molecule, which allows for complete customization.
Input
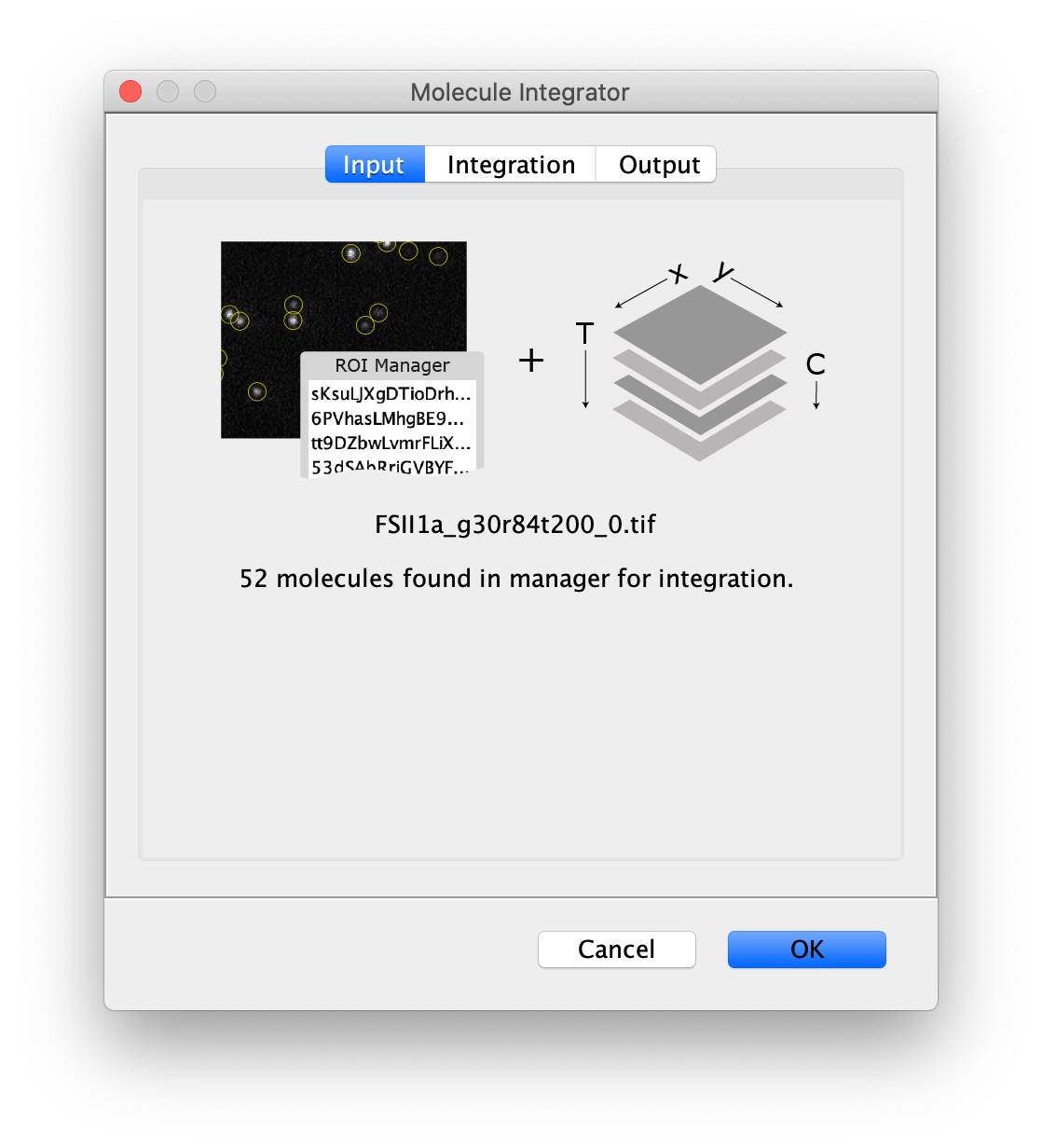
- Image - The selected image will by analyzed using the peaks in the ROI Manager. The name of the image is displayed in the dialog.
- ROI Manager - A list of point or circle ROIs that should be integrated with unique names need to be provided in the ROI Manager.
Integration
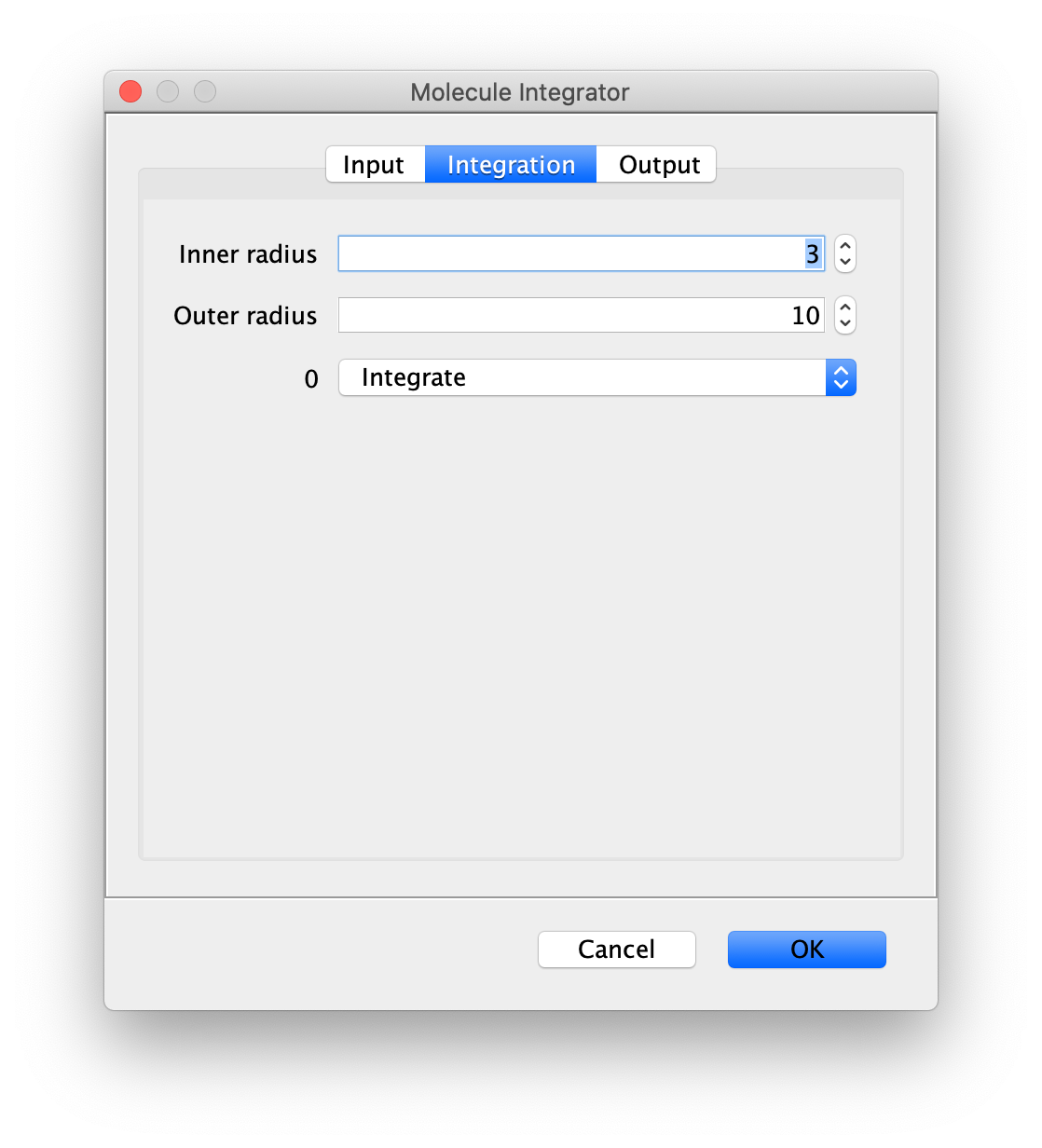
- Inner Radius - The radius of pixels around the peak to integrate. 0 means only one pixel, 1 means a radius of out beyond the center pixel and etc.
- Outer Radius - The radius of pixels around the Inner Radius to use for background correction. If the Inner Radius is 1 and the Outer Radius is 5. Then the circular region between 1 and 5 will be integrated and serve as the local background to correct the inner region.
- Channel Integration options - The options ‘Integrate’ and ‘Do not integrate’ are offered for all channels discovered in the input image. Channel names are used when provided. Otherwise, channel index starting from 0 is used.
Output
- Microscope - The microscope name added to the metadata record.
- Metadata UID - Options to either generate a metadata UID based on the metadata UID supplied in the image metadata or a new one that is random.
- Thread count - Determines how much computing power of your computer will be devoted to this calculation. A higher thread count decreases computing time.
- Verbose - This option includes extra table columns with more integration information. By default, the integrated peak signal corrected for median background and the median background used for the correction are included in the table of single molecule records generated. With the Verbose option checked, the table will also include the uncorrected integrated intensity and mean background.
The median background and mean background columns represent an estimate of the background signal within the inner radius region. First the median or mean pixel values in the outer region are calculated and then multiplied by the number of pixels in the inner region to represent an accurate estimate of the background signal in the inner region.
Result
- MoleculeArchive - A Single Molecule Archive in which each molecule record has the integrated fluorescence for each channel specified. Additionally, the peak position is saved for each time point (T) and Time (s) information is added if discovered.